Scientists just discovered a new way cells control their genes — it's called 'backtracking'
Scientists have discovered that, when a DNA-reading enzyme moves backwards along a gene, it may do so to help control when the gene is turned on.
The human body's roughly 30 trillion cells don't need all of their genes switched on at once. Instead, cells tightly control the activity of their genes — and recently, scientists uncovered a previously unknown way they accomplish that feat.
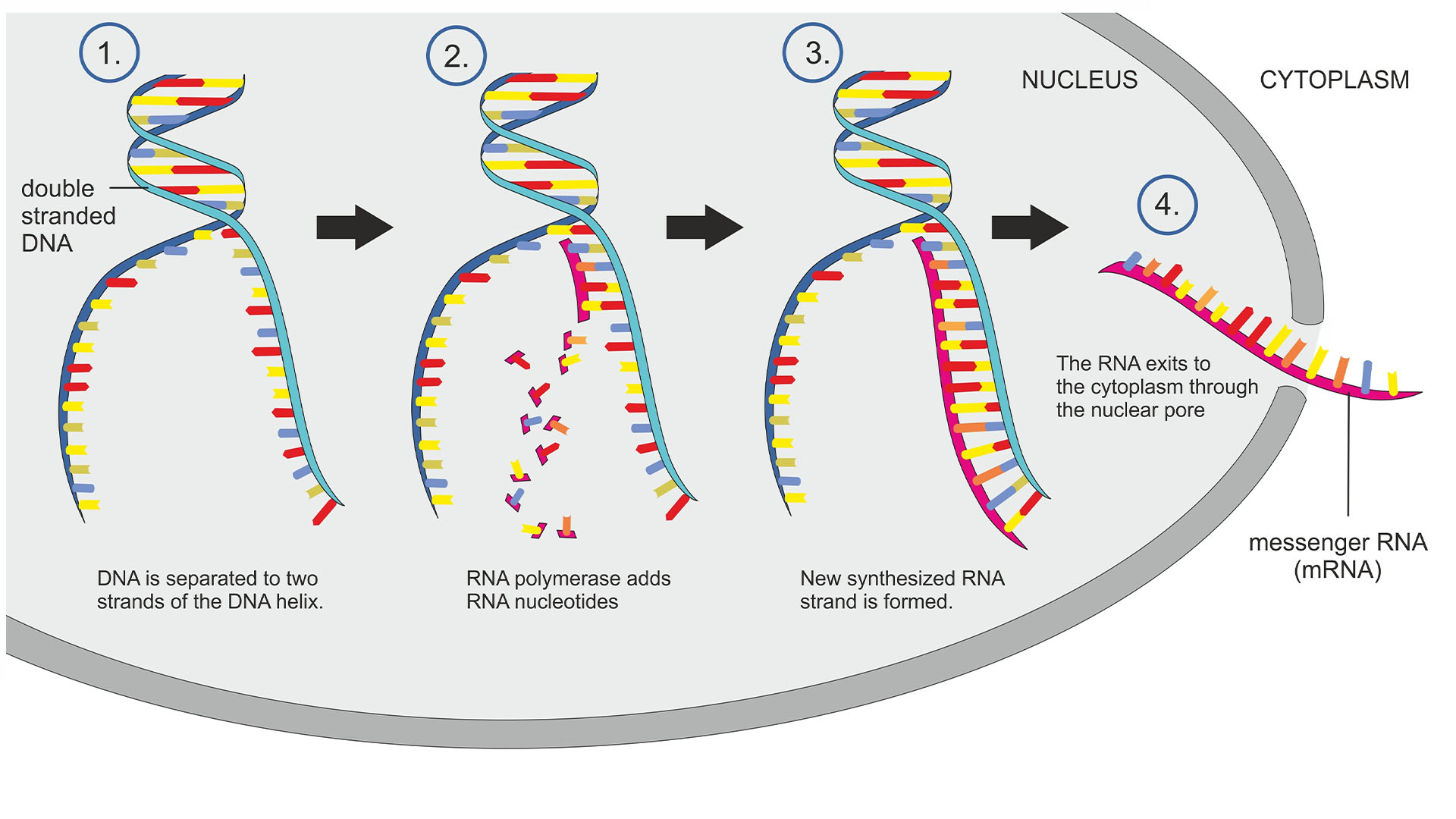
Human DNA contains approximately 20,000 to 25,000 genes. For a cell to function properly, the genetic code in that DNA is copied down, or transcribed, by an enzyme called RNA polymerase to make a molecule called RNA. Often, the RNA is then translated into proteins, the building blocks of life. There are myriad factors that determine which genes need to be turned on, such as the type of cell and its stage of development.
The recent study, published in February in the journal Molecular Cell, describes a newfound way by which cells control their genes. Called backtracking, it was initially thought to be a response to breaks in the DNA, but it's now being studied for its role in gene regulation.
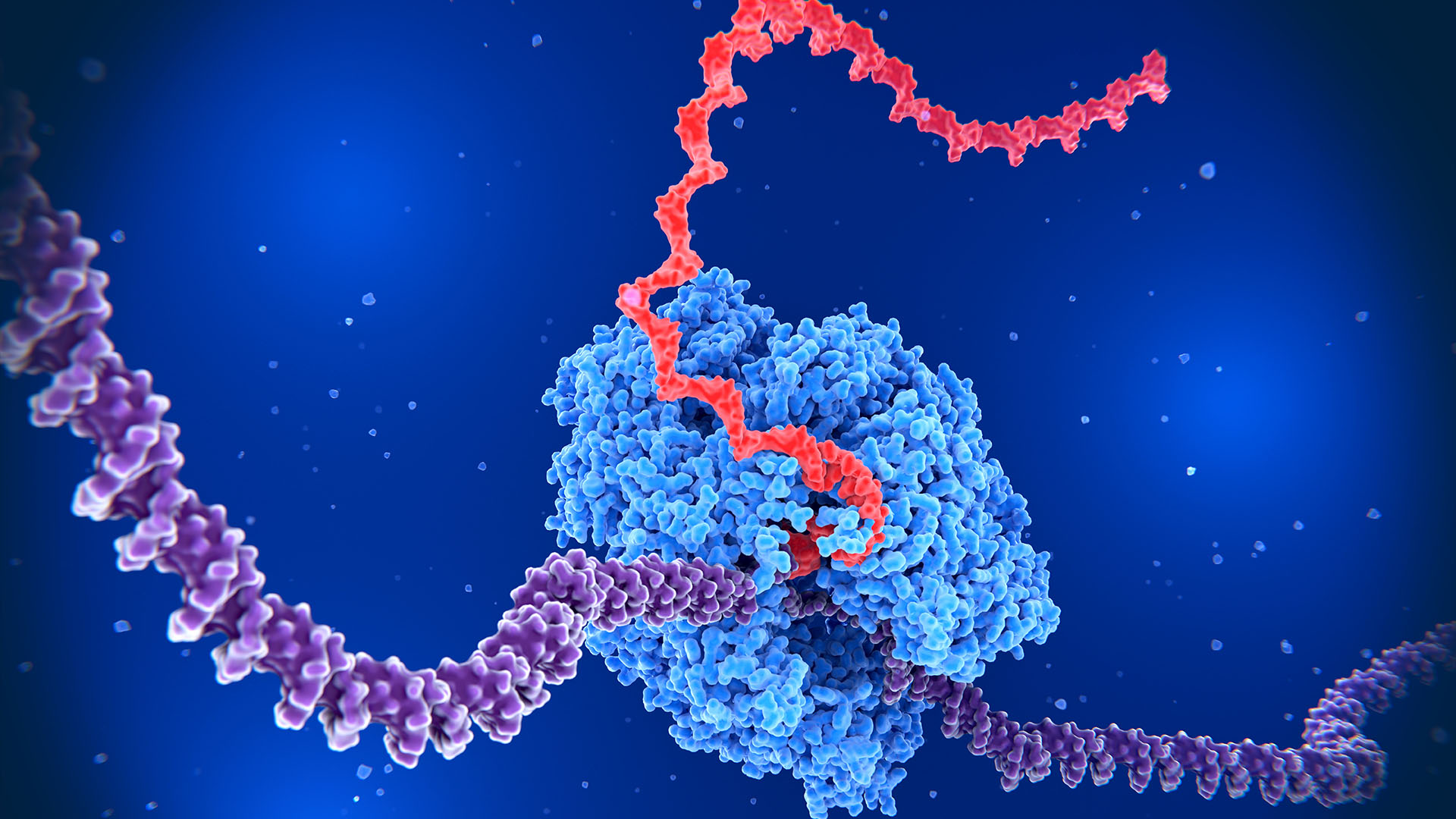
Discovered in 1997, backtracking is a process in which RNA polymerase, instead of moving forward along the DNA as it reads a gene, shifts back and pauses. This halt then resolves and the enzyme can move ahead again, churning out RNA.
Related: Scientists uncover hidden math that governs genetic mutations
"In early days, people thought that once RNA polymerase begins transcription, it will finish it without any problems," Evgeny Nudler, a professor of biochemistry at NYU Langone Health, told Live Science. "However, over the years, they realized that the picture is much more complicated." Nudler and colleagues published that first paper about backtracking in 1997.
When RNA polymerase backtracks a short distance, it extrudes a strand of newly formed RNA, causing the transcription process to pause. This extruded RNA is typically chopped off by enzymes, leaving the path clear for RNA polymerase to continue forward again.
Get the world’s most fascinating discoveries delivered straight to your inbox.
However, sometimes, the polymerase moves back a longer distance, and the extruded RNA blocks the site where those chopping enzymes usually snip. With this roadblock in place, the polymerase stays stuck in its backtracked state for longer periods, instead of just pausing for a short while.
Kevin Yang, a doctoral student in the Nudler Lab, and colleagues developed a new technique to capture the RNA strands extruded in persistent backtracking. The technique — called long-range cleavage sequencing, or LORAX-seq — reads the RNA's code to determine which genes are prone to this apparent hiccup in transcription. They developed this method to better detect backtracking events, and they were able to pick up thousands that would have been missed by previous methods.
"They have a very elegant method that very specifically pick[s] out and identifies cases where there's been long backtracking," David Bentley, a professor at the University of Colorado who was not involved in the study, told Live Science. "So even if they're very rare, they've got a very powerful method to pull them out."
"For the first time, we systematically mapped backtracked events," Nudler told Live Science. "And not just any backtracking events, but those which were extensive, where polymerase backtracks for long distances, gets stuck for a long time."
Related: Humans' big-brain genes may have come from 'junk DNA'
While the team expected to find backtracked events, they did not anticipate how prevalent they would be. They observed backtracking in many genes involved in making proteins from RNA; regulating cell division; and copying and packaging DNA.
The hotspot for persistent backtracking cropped up near gene promoters, which are the places where the RNA polymerase starts transcription, and splice sites, where RNA is trimmed to remove parts not needed for making proteins.
But what, exactly, is the role of backtracking? The researchers propose an intriguing theory: Backtracking may help RNA polymerase, along with hundreds of other proteins needed for gene transcription, "pause" at promoters and then begin pumping out RNAs as soon as the need arises. In processes such as cell division, during which massive amounts of proteins are needed, backtracking could allow for rapid activation of genes at a moment’s notice.
However, Bentley noted that "the relationship between backtracking and splicing is, I would say, still unresolved." So it's unclear if backtracking has a role at the splice sites.
The researchers also observed that the genes encoding histones — proteins that DNA winds around like a spool — are very prone to backtracking. These genes need to be very active after DNA copies itself during cell division, and so this backtracking may help time their activation at specific moments in the process.
With LORAX-seq established as a new way to detect backtracking, the method can now be used to study the role that this type of gene regulation plays in human diseases such as cancer, aging, and more broadly, any process that puts cells under stress.
Ever wonder why some people build muscle more easily than others or why freckles come out in the sun? Send us your questions about how the human body works to community@livescience.com with the subject line "Health Desk Q," and you may see your question answered on the website!

Sahana Sitaraman is a science writer based in Lausanne, Switzerland, specializing in biology. She particularly enjoys writing about unusual animal behaviours and the neuroscience behind them, mental health and women in STEM. She also dabbles in illustrating cool findings that pique her interest. In her free time, Sahana can be found out on a hike, acting it up with the local improv group or painting. She holds a bachelor's degree in microbiology from the University of Delhi, India and a master's and PhD in life sciences from the National Centre for Biological Sciences in Bangalore, India.
 Live Science Plus
Live Science Plus