Superbugs evolve inside the human body — tracking them in real time could help save patients, scientists say
A new proof-of-concept study explored the feasibility of tracking the evolution of superbug infections in real time to help save infected patients.
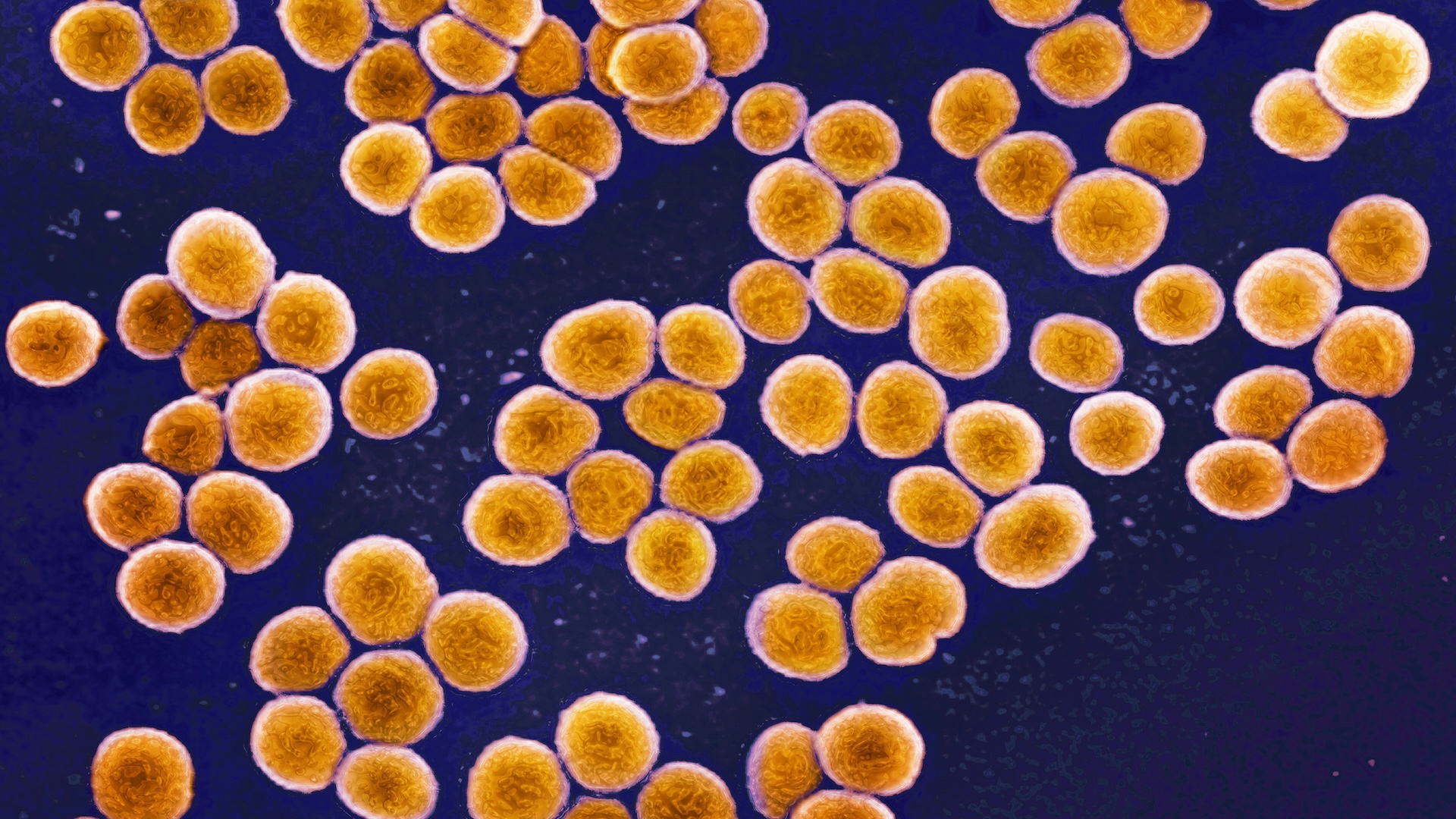
Bacterial "superbugs" can gain resistance to antibiotics as the microbes are actively infecting a person. Now, a new study suggests that treatments for these serious infections could be improved by tracking these genetic changes as they unfold in the bacteria.
"Our study is the first to show that by tracking bacterial evolution in real-time, genome sequencing can reveal tricks bacteria use to survive, giving doctors the power to stay one step ahead and tailor treatment to the specific bacterial strain," study co-author Dr. Stefano Giulieri, a clinician researcher and infectious-disease physician at the Doherty Institute in Melbourne, Australia, said in a statement.
The study, published in May in the journal Nature Communications, focused on Staphylococcus aureus. This bacterium is carried by about 30% of people and often causes no harm, but if it overgrows and triggers an infection, it can become resistant to antibiotic treatments. When bacteria are resistant to several antibiotics, they are considered dangerous superbugs.
To successfully treat a superbug infection, it's helpful to know if it's a "persistent" or "recurrent" infection. In a persistent infection, a patient continues testing positive even after five or more days of treatment. In a recurrent infection, the patient initially responds well to treatment but later tests positive for bacteria again, either with the same strain or a new type. Understanding the type of infection can help steer treatment by guiding doctors' choice of drugs.
To see if genetic analyses could help guide these decisions, the researchers analyzed S. aureus samples from 11 patients whose antibiotic treatments were failing. This included 60 different strains of the bacterium.
Related: How fast can antibiotic resistance evolve?
Using genetic analyses, the research team identified if each patient's samples contained bacteria of the same strains or from genetically distinct strains. Next, they ran tests to spot signatures of adaptive evolution, meaning signs the microbes were picking up traits that would help them to survive better. Adaptive evolution enables bacteria to survive even with antibiotics present.
Get the world’s most fascinating discoveries delivered straight to your inbox.
About one-third of the sampled strains showed signs of adaptive evolution, specifically in genes previously linked to antibiotic resistance. This suggested that the patients' antibiotic treatments should be switched to a medication that the bacteria weren't resistant to.
But a question remained: Would this information actually be helpful to doctors as they were treating superbug infections?
To investigate, the scientists created a survey based on the 11 patients' cases, including descriptions with and without the evolution analysis. They recruited 25 infectious-disease doctors from around the world to answer the survey. When given the evolution report, 34% of the physicians changed their original antibiotic regimen suggestions, switching antibiotics and/or adjusting the duration patients stayed on the same drug.
These findings hint that, in actual clinical practice, tracking bacterial evolution could improve doctors' assessments of antibiotic failure and subsequent treatment decisions.
Although the new study has some limitations, such as its small sample size, it provides a "proof of concept" for using evolutionary analyses as a tool for combating antibiotic-resistant infections, the study authors wrote in their report.
"This tool can significantly impact our decision making process," said Dr. Quyen Nguyen, an assistant professor of medicine at the University of Pittsburgh who was not involved in the research. "Therefore, we welcome new technology that can quickly give more precise data so that we can increase confidence in our decisions," Nguyen told Live Science in an email.
Currently, the cost and turnaround time of genomic sequencing are still hurdles to using this approach regularly with patients. Future studies will need to address how the framework could best be applied and explore its use in larger groups of patients, the study authors concluded.
This article is for informational purposes only and is not meant to offer medical advice.

Shira Gordon is a freelance health and science writer. She has a PhD from the University of Cincinnati in biology. She spent almost a decade in the lab after her doctorate degree. Her research focused on animal communication, blending neurophysiology, behavior, and biomechanics. Now as a science communicator, she covers human health, animals, and ecology. In addition to writing articles, Gordon has worked on website content for numerous NIH and HHS institutes and has produced and written scripts for award-winning videos.
You must confirm your public display name before commenting
Please logout and then login again, you will then be prompted to enter your display name.
 Live Science Plus
Live Science Plus





