Meet 'Fanzor,' the 1st CRISPR-like system found in complex life
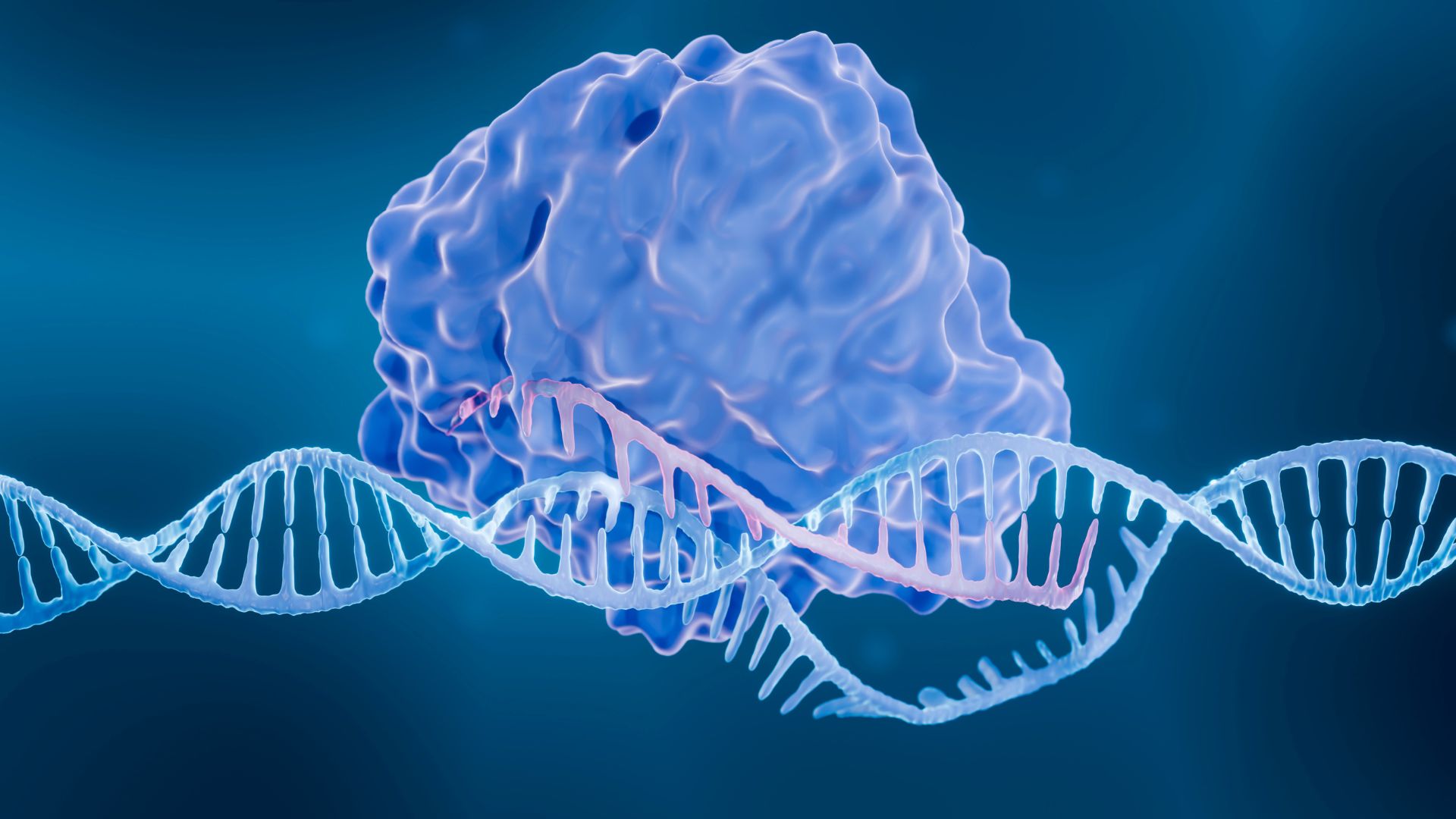
Scientists discovered Fanzor proteins, which work like CRISPR but are smaller and more easily delivered into cells, and used them to edit human DNA.
Get the world’s most fascinating discoveries delivered straight to your inbox.
You are now subscribed
Your newsletter sign-up was successful
Want to add more newsletters?

Delivered Daily
Daily Newsletter
Sign up for the latest discoveries, groundbreaking research and fascinating breakthroughs that impact you and the wider world direct to your inbox.

Once a week
Life's Little Mysteries
Feed your curiosity with an exclusive mystery every week, solved with science and delivered direct to your inbox before it's seen anywhere else.

Once a week
How It Works
Sign up to our free science & technology newsletter for your weekly fix of fascinating articles, quick quizzes, amazing images, and more

Delivered daily
Space.com Newsletter
Breaking space news, the latest updates on rocket launches, skywatching events and more!

Once a month
Watch This Space
Sign up to our monthly entertainment newsletter to keep up with all our coverage of the latest sci-fi and space movies, tv shows, games and books.

Once a week
Night Sky This Week
Discover this week's must-see night sky events, moon phases, and stunning astrophotos. Sign up for our skywatching newsletter and explore the universe with us!
Join the club
Get full access to premium articles, exclusive features and a growing list of member rewards.
Researchers have identified a new gene-editing system similar to CRISPR in complex organisms, demonstrating for the first time that DNA-modifying proteins exist across all kingdoms of life.
Feng Zhang, a biochemist at the Broad Institute of MIT and Harvard and the McGovern Institute for Brain Research at MIT, led the team and previously co-discovered the gene-editing potential of the CRISPR-Cas9 system, which functions as a kind of "molecular scissors" that remove sections of DNA, thus disabling genes or allowing new ones to be swapped in.
Prior to this discovery, such systems had only been found in simple organisms such as bacteria and archaea, which wield them as a sort of rudimentary immune system for chopping up the DNA of invaders. Researchers detected the newfound system, called Fanzor, in fungi, algae, amoebas and a species of clam, vastly broadening the groups known to use these genetic tools.
"People have been saying with such certainty for so long that eukaryotes [organisms whose complex cells contain nuclei] couldn't have a similar system," said Ethan Bier, a geneticist at the University of California San Diego, who uses gene editing in his work but was not involved in the study. "But it's typical cleverness from the Zhang lab, proving them wrong," Bier told Live Science.
Related: CRISPR-edited fat shrank tumors in mice. Someday, it could work in people, scientists say.
After publishing their first paper on CRISPR in 2013, Zhang and colleagues began studying how these systems evolve. During this work, the group identified a class of proteins in bacteria called OMEGAs, thought to be early ancestors of Cas9 proteins, the "scissors" of the CRISPR system. They began to suspect that Fanzor proteins, a type of OMEGA, could also be modifying DNA.
The group screened online databases for the proteins and were surprised to find several in samples isolated from fungi, protists, arthropods, plants and giant viruses. The thinking, Zhang said, is that the genes needed to make Fanzor proteins got shuffled from bacteria into complex organisms through a process known as horizontal gene transfer. Genes that encode for Fanzor proteins were integrated into the genomes of eukaryotic organisms within transposable elements, meaning bits of DNA that can move about the genome and replicate themselves.
Get the world’s most fascinating discoveries delivered straight to your inbox.
In experiments, the researchers found that Fanzor proteins share some similarities with CRISPR. Fanzor proteins also interact with guide RNA, a molecule that guides the proteins to the DNA destined to be cut. This molecule, called an omegaRNA, complements the strand of target DNA. When they match up, the two pieces zip together and Fanzor can then cut the DNA.
The team tested the Fanzor system in human cells but at first found that it was relatively inefficient at adding or removing bits of DNA, completing the process successfully about 12% of the time. After some creative engineering to enhance and stabilize the system, however, the researchers bumped the efficiency up to just over 18%.
This inefficiency isn't surprising, according to Bier, nor a sign that Fanzor isn't as good as CRISPR. Scientists have engineered CRISPR so that it can make the desired substitutions almost every time, but "it certainly didn’t start out that way," he said. But Bier added it will be hard for Fanzor to match Cas9, which he called "the most adaptable and forgiving protein for the types of things you want to do to it."
Fanzor will instead likely complement CRISPR, which has been used both in research and in experimental medical treatments for conditions like blindness and cancer.
Compared with CRISPR, "the Fanzor systems are more compact and therefore have the potential to be more easily delivered to cells and tissues," Zhang said, and they're less prone to accidentally degrading nearby RNA or DNA — so-called off-target or collateral effects. This makes Fanzor attractive for use in gene therapy.
Zhang told Live Science he's now excited to go looking for similar systems in new places.
"This work really underscores the power of studying biodiversity," Zhang said. "There are likely more RNA-guided systems out there in nature that hold future promise for gene editing."

Amanda Heidt is a Utah-based freelance journalist and editor with an omnivorous appetite for anything science, from ecology and biotech to health and history. Her work has appeared in Nature, Science and National Geographic, among other publications, and she was previously an associate editor at The Scientist. Amanda currently serves on the board for the National Association of Science Writers and graduated from Moss Landing Marine Laboratories with a master's degree in marine science and from the University of California, Santa Cruz, with a master's degree in science communication.
 Live Science Plus
Live Science Plus










