New Gene Sequencing Method Could Reduce Cost, Increase Speed
Get the world’s most fascinating discoveries delivered straight to your inbox.
You are now subscribed
Your newsletter sign-up was successful
Want to add more newsletters?

Delivered Daily
Daily Newsletter
Sign up for the latest discoveries, groundbreaking research and fascinating breakthroughs that impact you and the wider world direct to your inbox.

Once a week
Life's Little Mysteries
Feed your curiosity with an exclusive mystery every week, solved with science and delivered direct to your inbox before it's seen anywhere else.

Once a week
How It Works
Sign up to our free science & technology newsletter for your weekly fix of fascinating articles, quick quizzes, amazing images, and more

Delivered daily
Space.com Newsletter
Breaking space news, the latest updates on rocket launches, skywatching events and more!

Once a month
Watch This Space
Sign up to our monthly entertainment newsletter to keep up with all our coverage of the latest sci-fi and space movies, tv shows, games and books.

Once a week
Night Sky This Week
Discover this week's must-see night sky events, moon phases, and stunning astrophotos. Sign up for our skywatching newsletter and explore the universe with us!
Join the club
Get full access to premium articles, exclusive features and a growing list of member rewards.
This Behind the Scenes article was provided to LiveScience in partnership with the National Science Foundation.
The first human genome took 13 years and $3 billion to produce. Today, geneticists can generate the same information in a matter of months, for a fraction of the cost.
As "next-generation" gene sequencers begin to make their mark on the life sciences, teams around the world are racing to develop new and improved DNA sequencers that can ingest a strand of nucleotide bases and directly "read" a person's genetic code for less than $1,000.
The medical community predicts that the advent of the $1000 personal genome will lead to major changes in the understanding and treatment of illness. Researchers will be able to perform widespread comparative studies to correlate disease to gene expression. Chemists will design genetically-targeted drugs, and doctors will deliver medical treatments based on a patient's specific genetic disposition.
Aleksei Aksimentiev, a computational physicist at the University of Illinois Urbana-Champaign (UIUC), is working to develop a new kind of sequencer that will make the dream of the $1000 genome a reality.
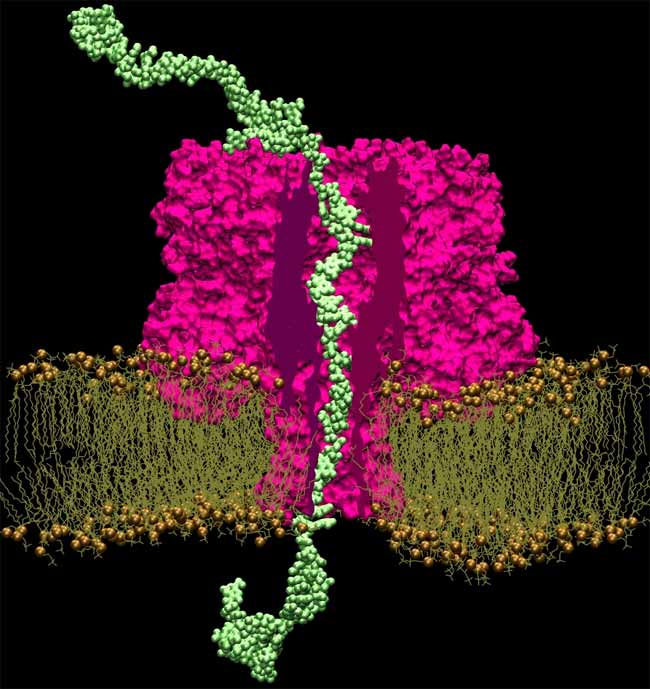
The sequencer uses an electric field to drive a strand of DNA through a small hole, or "nanopore," in a membrane. If this process can be controlled, the sequencer will be able to read base pairs in order by measuring the change in current as each pair moves through the hole in the membrane.
A great concept — but does it work?
Get the world’s most fascinating discoveries delivered straight to your inbox.
Not yet, according to Aksimentiev. The experimental designs of these sequencers are plagued by noisy signals, blockages and DNA base pairs that pass too quickly through the pore.
"This intuitively simple physical picture doesn't apply in practice," Aksimentiev said.
Since experiments can't show exactly what's going on inside a nanopore, Aksimentiev produces atom-by-atom models of nanopore designs and set them in motion using the National Science Foundation-sponsored Ranger supercomputer at the Texas Advanced Computing Center (TACC). His simulations revealed the atomic-level movements of DNA as it wriggled through a nanopore, leading to insights into how to improve the design of the system.
"The simulations explain what processes give rise to the signals we measure," he said. Aksimentiev discovered that the key requirement for sequencing DNA directly is to keep the strand in the pore long enough to read out its sequence.
"If the DNA moves too fast, then one cannot read out the signal to distinguish the difference between the base pairs," he said. "We have to find a way to trap the DNA."
Aksimentiev and his colleagues believe they've found a way.
When DNA comes in contact with an electrical field, it stretches. Applying that field on one side of the membrane forces the strands into a pore smaller than it would usually fit in. Turning off the field, on the other hand, causes the DNA to relax and traps the molecule in the hole. By pulsing the field, stretching and relaxing the DNA, the strand moves, base-by-base, through the pore, at a speed slow enough to be read clearly.
The team filed for a provisional patent in 2009 for their design of the first nanopore sequencer able to accommodate double-stranded DNA.{{ video="LS_100709_dna-nanopore" title="New Gene Sequencing Tool Moves DNA With Electric Field" caption="Researchers are developing a new method to sequence the human genome. The sequencer uses an electric field to drive a strand of DNA through a small hole, or "nanopore," in a membrane. Credit: Aleksei Aksimentiev, University of Illinois at Urbana-Champaign" }}
High-performance computing systems are required for this kind of research because of the precision needed to differentiate between the four DNA nucleotides, which differ by only a few atoms.
"Dr. Aksimentiev's simulations represent our eyes," said Greg Timp, Aksimentiev's collaborator and professor of electrical engineering at the University of Notre Dame. "We can't see without them."
Aksimentiev's improved nanopore designs promise a dramatic reduction in the cost of gene sequencing, and a commensurate increase in speed. The development of such a sequencer would have important ramifications for medicine, biology and human health.
"It will have a noticeable impact on the way we understand and treat human disease," Aksimentiev said. "Everyone would be able to afford their DNA sequence."
If this sounds like science fiction, you won't have to wait long to find out how it turns out. The National Institute of Health (NIH) has set a goal of producing a $1,000 genome using nanopore sequencers by 2013.
Soon, we'll all have access to our genetic information. What we do with this knowledge remains to be seen.
This work is supported by grants from NIH's "Revolutionary Genome Sequencing Technologies" program, and NSF's "Physics Frontier Center" grant in support the Center for the Physics of Living Cells.
- 10 Profound Innovations Ahead
- The Top 10 Worst Hereditary Conditions
- Scrutiny of Personal Gene Tests Increases
Editor's Note: This research was supported by the National Science Foundation (NSF), the federal agency charged with funding basic research and education across all fields of science and engineering. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author and do not necessarily reflect the views of the National Science Foundation. See the Behind the Scenes Archive.
 Live Science Plus
Live Science Plus










