Cells Shimmer Like a Thousand Ice Cream Sprinkles in Gorgeous New 'DNA Microscope' Images
Get the world’s most fascinating discoveries delivered straight to your inbox.
You are now subscribed
Your newsletter sign-up was successful
Want to add more newsletters?

Delivered Daily
Daily Newsletter
Sign up for the latest discoveries, groundbreaking research and fascinating breakthroughs that impact you and the wider world direct to your inbox.

Once a week
Life's Little Mysteries
Feed your curiosity with an exclusive mystery every week, solved with science and delivered direct to your inbox before it's seen anywhere else.

Once a week
How It Works
Sign up to our free science & technology newsletter for your weekly fix of fascinating articles, quick quizzes, amazing images, and more

Delivered daily
Space.com Newsletter
Breaking space news, the latest updates on rocket launches, skywatching events and more!

Once a month
Watch This Space
Sign up to our monthly entertainment newsletter to keep up with all our coverage of the latest sci-fi and space movies, tv shows, games and books.

Once a week
Night Sky This Week
Discover this week's must-see night sky events, moon phases, and stunning astrophotos. Sign up for our skywatching newsletter and explore the universe with us!
Join the club
Get full access to premium articles, exclusive features and a growing list of member rewards.
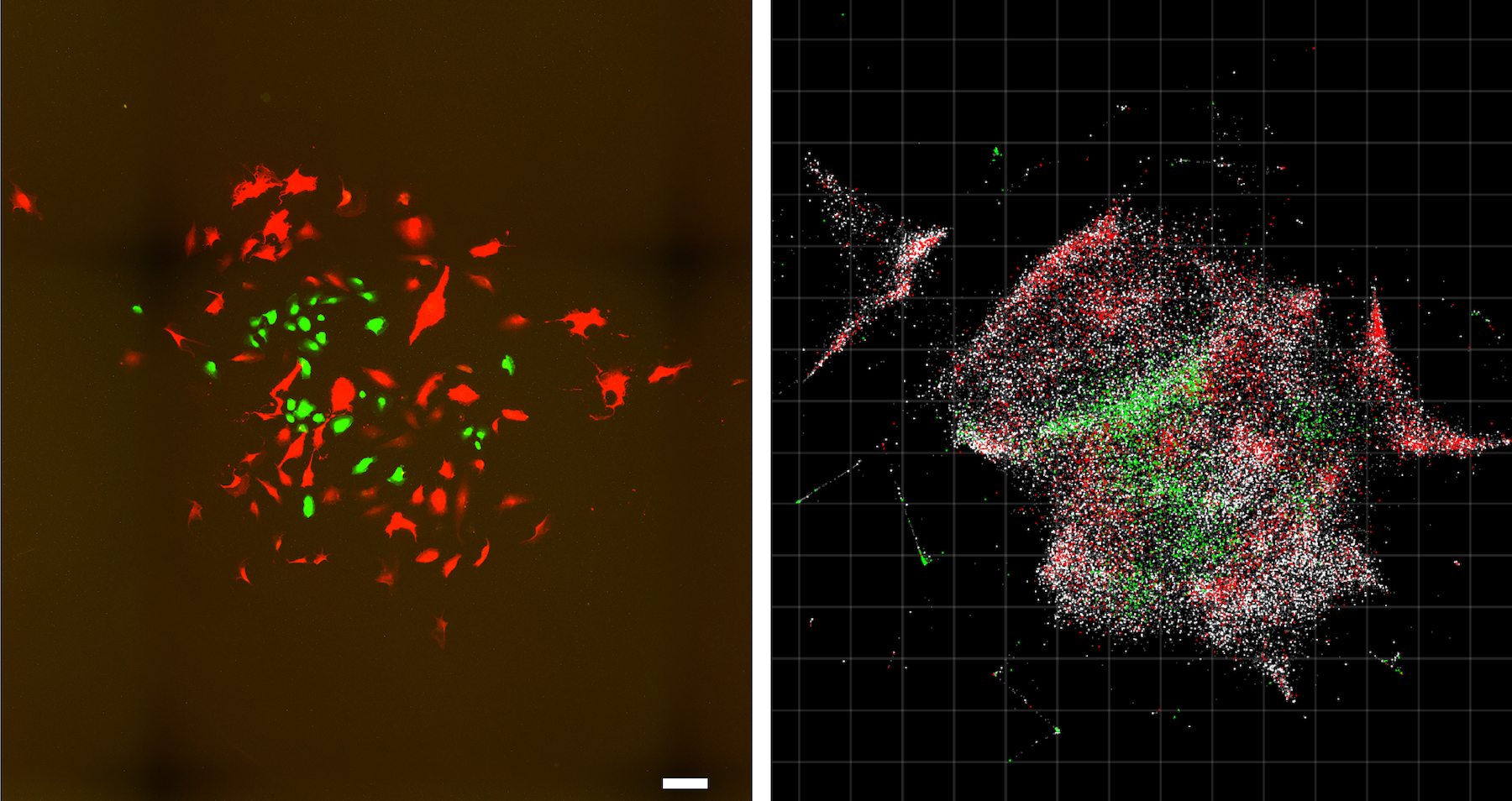
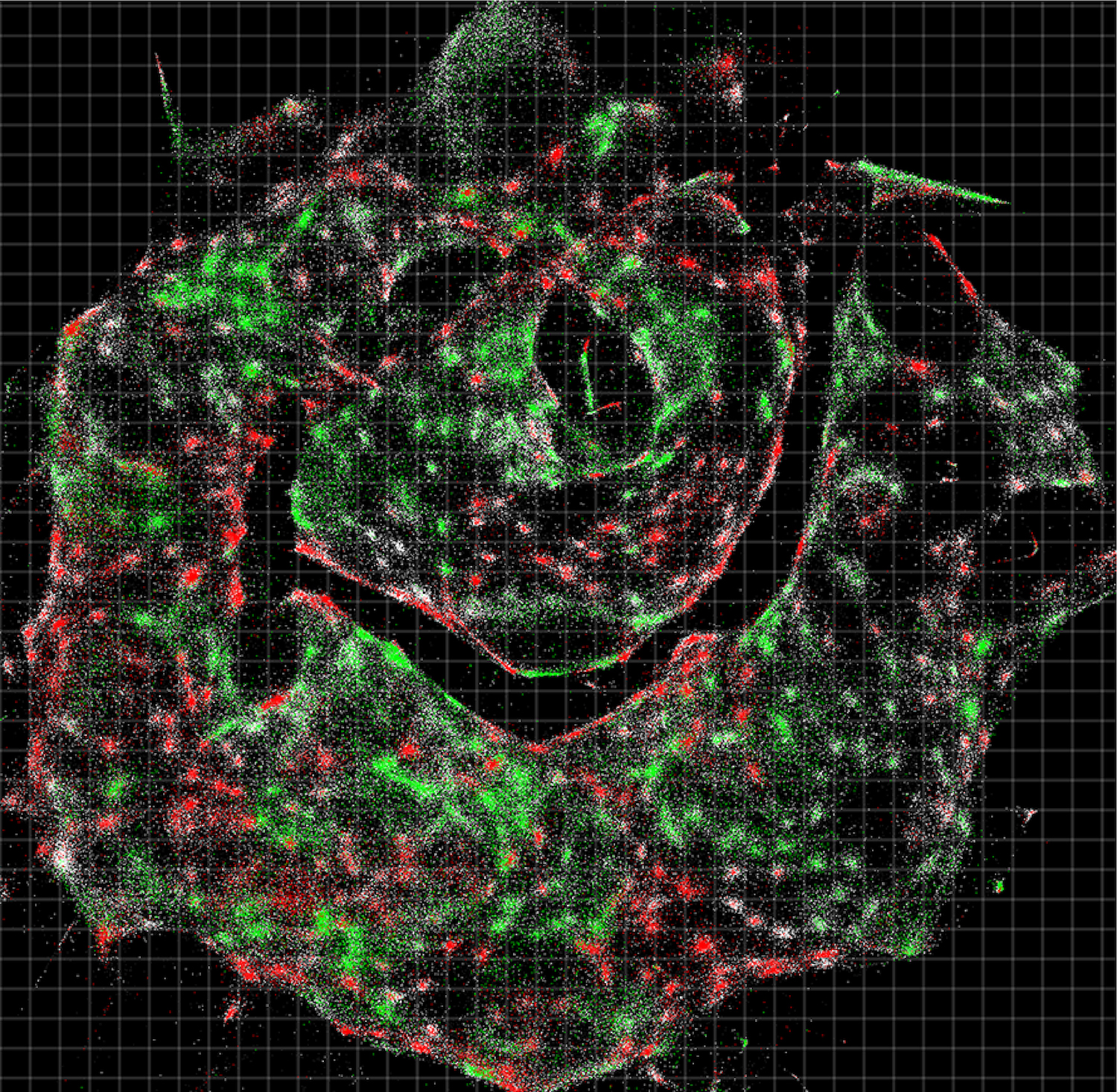
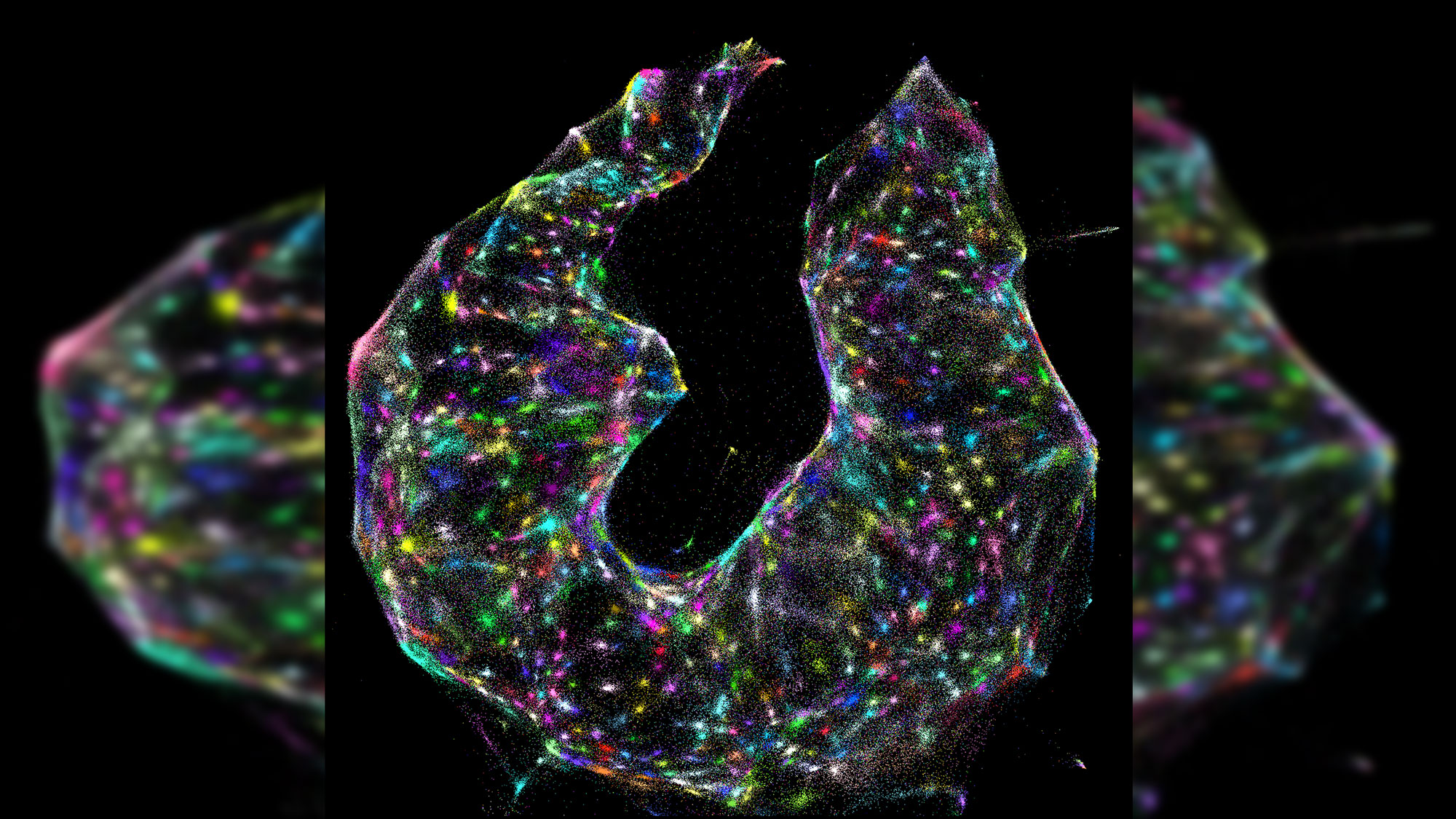
What looks like a kaleidoscope of glowing ice cream sprinkles or a cross between a nebula and a 1980s dance party is actually something even more astonishing: an unfettered and detailed view of the exact locations of DNA and RNA inside a living cell.
The method that opened the doors for this unprecedented look inside living cells — known as DNA microscopy — was perfected over a period of six years, according to a new study.
"DNA microscopy is an entirely new way of visualizing cells that captures both spatial and genetic information simultaneously from a single specimen," study lead researcher Joshua Weinstein, a postdoctoral associate at the Broad Institute of MIT, said in a statement. [Check Out These Amazing Super-Detailed Images of Fruit Fly Brains]
The technique even allows researchers to see the exact order of nucleotides, the "letters" that make up DNA's double helix and RNA's single strand, within each cell.
"It will allow us to see how genetically unique cells — those comprising the immune system, cancer or the gut, for instance — interact with one another and give rise to complex multicellular life," Weinstein said.
Over the past few decades, researchers have developed myriad tools that help them collect molecular data from tissue samples. But efforts to pair this technology with spatial data — so that researchers know where and how genetic material inside a cell is arranged — often involves expensive and specialized machinery.
The new approach makes the process much easier, the researchers said. In essence, the method uses tiny tags — made out of customized DNA sequences each about 30 nucleotides long — that latch onto every DNA and RNA molecule in a cell. Then, the tags are replicated until there are hundreds of copies of them within the cell. As these copies interact with one another, they combine and make unique DNA labels, the researchers said.
Get the world’s most fascinating discoveries delivered straight to your inbox.
The interactions between these DNA tags is key. Once researchers collect the labeled biomolecules and sequence them, they can use a computer algorithm to decode and reconstruct the tags' original positions in the cell, creating a color-coded virtual image of the sample. Pinpointing the location of each molecule is similar to how cellphone towers triangulate the locations of nearby cellphones, the researchers said.
The technique may help researchers better understand different kinds of human disease. For instance, in the study the researchers showed that DNA microscopy could map the locations of individual human cancer cells in a sample. These synthetic DNA tags can even help scientists map the locations of antibodies, receptors and molecules on tumor cells, they said.
"We've used DNA in a way that's mathematically similar to photons in light microscopy," Weinstein said. "This allows us to visualize biology as cells see it and not as the human eye does."
The study was published online yesterday (June 20) in the journal Cell.
- Tiny & Nasty: Images of Things That Make Us Sick
- Images: Human Parasites Under the Microscope
- Photos: Amazing Microscopic Views of Italian Cocktails
Originally published on Live Science.

Laura is the managing editor at Live Science. She also runs the archaeology section and the Life's Little Mysteries series. Her work has appeared in The New York Times, Scholastic, Popular Science and Spectrum, a site on autism research. She has won multiple awards from the Society of Professional Journalists and the Washington Newspaper Publishers Association for her reporting at a weekly newspaper near Seattle. Laura holds a bachelor's degree in English literature and psychology from Washington University in St. Louis and a master's degree in science writing from NYU.
 Live Science Plus
Live Science Plus