Coronavirus hijacks cells, forces them to grow tentacles, then invades others
Scientists are looking at how the coronavirus invades cells to find the right drugs to target it.

Get the world’s most fascinating discoveries delivered straight to your inbox.
You are now subscribed
Your newsletter sign-up was successful
Want to add more newsletters?

Delivered Daily
Daily Newsletter
Sign up for the latest discoveries, groundbreaking research and fascinating breakthroughs that impact you and the wider world direct to your inbox.

Once a week
Life's Little Mysteries
Feed your curiosity with an exclusive mystery every week, solved with science and delivered direct to your inbox before it's seen anywhere else.

Once a week
How It Works
Sign up to our free science & technology newsletter for your weekly fix of fascinating articles, quick quizzes, amazing images, and more

Delivered daily
Space.com Newsletter
Breaking space news, the latest updates on rocket launches, skywatching events and more!

Once a month
Watch This Space
Sign up to our monthly entertainment newsletter to keep up with all our coverage of the latest sci-fi and space movies, tv shows, games and books.

Once a week
Night Sky This Week
Discover this week's must-see night sky events, moon phases, and stunning astrophotos. Sign up for our skywatching newsletter and explore the universe with us!
Join the club
Get full access to premium articles, exclusive features and a growing list of member rewards.
Cells infected with the new coronavirus grow stringy, tentacle-like arms that allow the virus to invade other cells, according to a new study.
The novel coronavirus, known as SARS-CoV-2, has now infected more than 12.2 million people worldwide and killed more than 555,500, according to the Johns Hopkins dashboard. To defeat the virus, researchers around the world are taking part in an unprecedented effort to find new drugs and repurpose old ones.
But to truly find the right therapeutic weapon, scientists need to understand in detail how the virus invades human cells. To figure that out, an international group of researchers looked at how the virus changes activity inside cells in order to invade more and more cells. They specifically analyzed how the virus can alter certain proteins in infected cells. (Proteins carry out the instructions of genes, and so protein changes could impact the actual actions of infected cells.)
Related: Coronavirus Live Updates
This new research builds upon a "blueprint" of 332 human proteins that interact with 27 SARS-CoV-2 viral proteins that researchers had described in April in the journal Nature. In this new study, the researchers analyzed all of the human proteins that, when infected, showed changes in a process called phosphorylation — in which a protein called a kinase sticks a phosphoryl group (a phosphorus atom attached to three oxygens) onto other proteins, according to a statement.
Phosphorylation, a process that can activate and deactivate proteins, is "extremely important" for many cellular processes, including protein synthesis, cell division, signaling, cell growth, development and aging, according to an article published in June 2017 in the International Journal of Molecular Medicine.
The set of kinases is like the "master switchboard of the cell," said senior author Nevan Krogan, the director of the Quantitative Biosciences Institute (QBI) at the School of Pharmacy at the University of California, San Francisco and senior investigator at Gladstone Institutes. "If the virus can come in and manipulate the switchboard, it can manipulate things in a way that's beneficial for infection."
Get the world’s most fascinating discoveries delivered straight to your inbox.
Using a method called mass spectrometry, which measures the mass of different molecules such as proteins, the team found "dramatic rewiring of phosphorylation on host and viral proteins," inside monkey cells, the authors wrote in their new study. This method can identify the subtle change in mass between a phosphorylated and a non-phosphorylated protein, Krogan told Live Science.
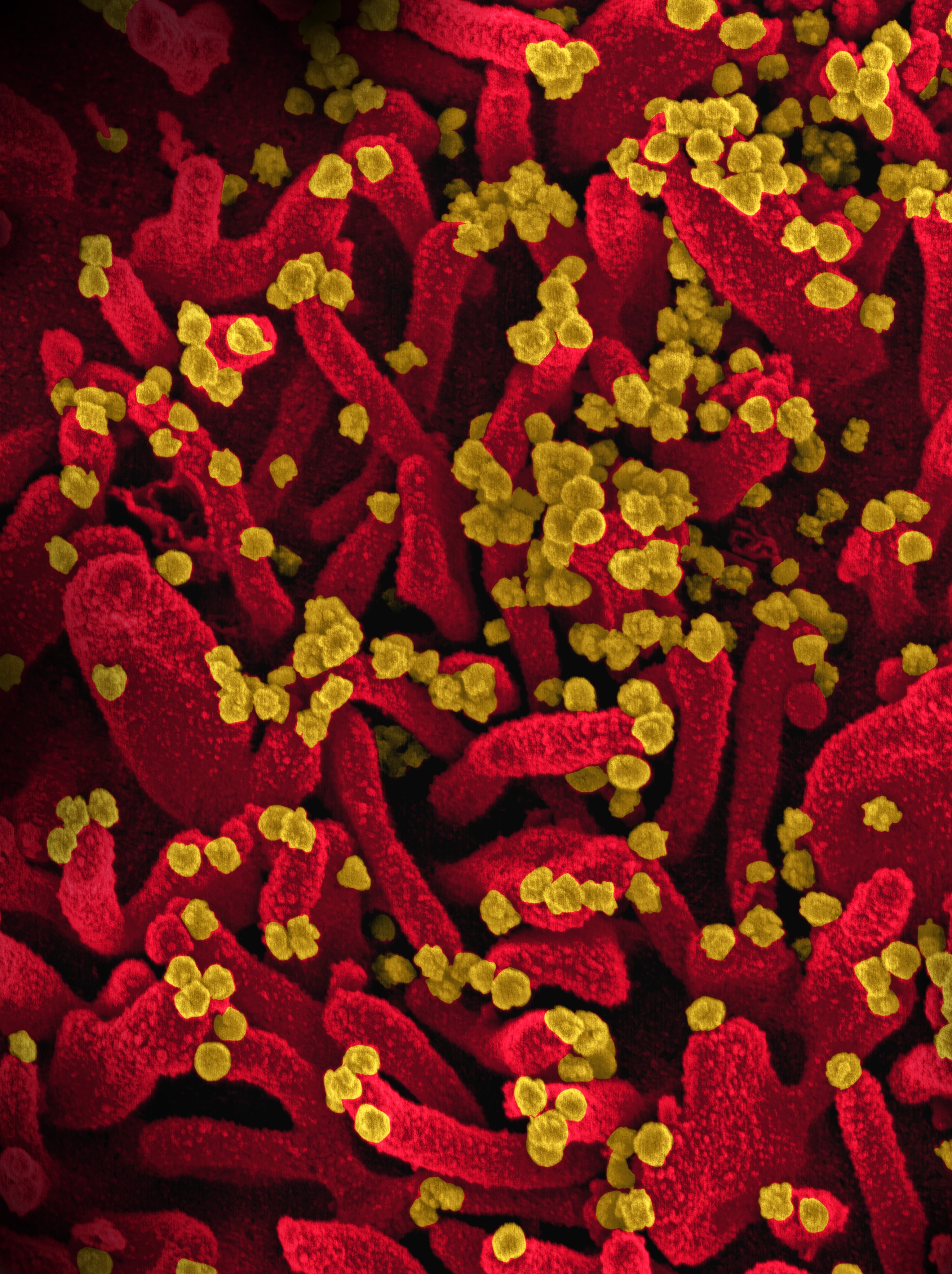
Stringy cells
Human cells have proteins that are very similar to those in monkey cells, Krogan said. The team found that 40 of the 332 human proteins that were previously found to interact with the coronavirus were phosphorylated differently in monkey cells infected with the virus compared with those not infected.
In addition, out of 518 kinases they examined, the scientists found 49 showed changes in phosphorylation activity, according to the statement. The activity of a couple of kinases, including what's called a casein kinase II (CK2), were dramatically altered by the virus and lie within important cell-signaling pathways, according to the statement.
Related: 20 of the worst epidemics and pandemics in history
High-resolution imaging of the infected cells showed that the cells had grown tentacle-like protrusions called "filopodia," which contained viral proteins, according to the statement. The researchers found both CK2 and viral proteins within the filopodia, suggesting that the coronavirus hijacks CK2 and forces it to form tentacles. Those tentacles then poke holes in nearby cells, allowing the virus to infect new cells, according to the statement.
The researchers then identified 87 drugs that are either approved by the U.S. Food and Drug Administration (FDA) or currently in clinical trials that might target some of these kinases or pathways that are altered by SARS-CoV-2 in both human and monkey cells. Kinases are "very druggable," Krogan said. They found that seven of these compounds, mostly anti-cancer and anti-inflammatory compounds, inhibited the virus from replicating and growing in both human lung cells and monkey kidney cells, Krogan said.
This study "is careful and thorough, but has some limitations," said Carol Shoshkes Reiss, a professor of Biology and Neural Science at New York University, who was not a part of the study. The researchers tested how the virus infects cells using non-human cells, rather than primary human airway cells, she said. The authors acknowledge this limitation in the study, she added. They also didn't demonstrate their ideas worked in any of the top animal models used to study SARS-CoV-2 infections, such as transgenic mice or hamsters, she told Live Science in an email.
"It definitely has potential, if tested appropriately," she said. "But, you have to realize that these pathways are essential, and although there are licensed drugs that target them, the potential for side effects and off-target impact is high."
The findings were published in the journal Cell on June 28. (The Krogan Lab received research support from Vir Biotechnology and F. Hoffmann-La Roche and one of the co-authors has consulting agreements with some pharmaceutical companies.)
Originally published on Live Science.

Yasemin is a staff writer at Live Science, covering health, neuroscience and biology. Her work has appeared in Scientific American, Science and the San Jose Mercury News. She has a bachelor's degree in biomedical engineering from the University of Connecticut and a graduate certificate in science communication from the University of California, Santa Cruz.
 Live Science Plus
Live Science Plus










