Spinning Samples Yield Insights into Disease and Harnessing Solar Power
Get the world’s most fascinating discoveries delivered straight to your inbox.
You are now subscribed
Your newsletter sign-up was successful
Want to add more newsletters?

Delivered Daily
Daily Newsletter
Sign up for the latest discoveries, groundbreaking research and fascinating breakthroughs that impact you and the wider world direct to your inbox.

Once a week
Life's Little Mysteries
Feed your curiosity with an exclusive mystery every week, solved with science and delivered direct to your inbox before it's seen anywhere else.

Once a week
How It Works
Sign up to our free science & technology newsletter for your weekly fix of fascinating articles, quick quizzes, amazing images, and more

Delivered daily
Space.com Newsletter
Breaking space news, the latest updates on rocket launches, skywatching events and more!

Once a month
Watch This Space
Sign up to our monthly entertainment newsletter to keep up with all our coverage of the latest sci-fi and space movies, tv shows, games and books.

Once a week
Night Sky This Week
Discover this week's must-see night sky events, moon phases, and stunning astrophotos. Sign up for our skywatching newsletter and explore the universe with us!
Join the club
Get full access to premium articles, exclusive features and a growing list of member rewards.
This Behind the Scenes article was provided to LiveScience in partnership with the National Science Foundation.
In 1926, Theodor Svedberg won the Nobel Prize in Chemistry for a novel method of separating proteins based on a device he invented: the analytic ultracentrifuge.This technique has yielded insights into the purity, structure and behavior of proteins, DNA and RNA.
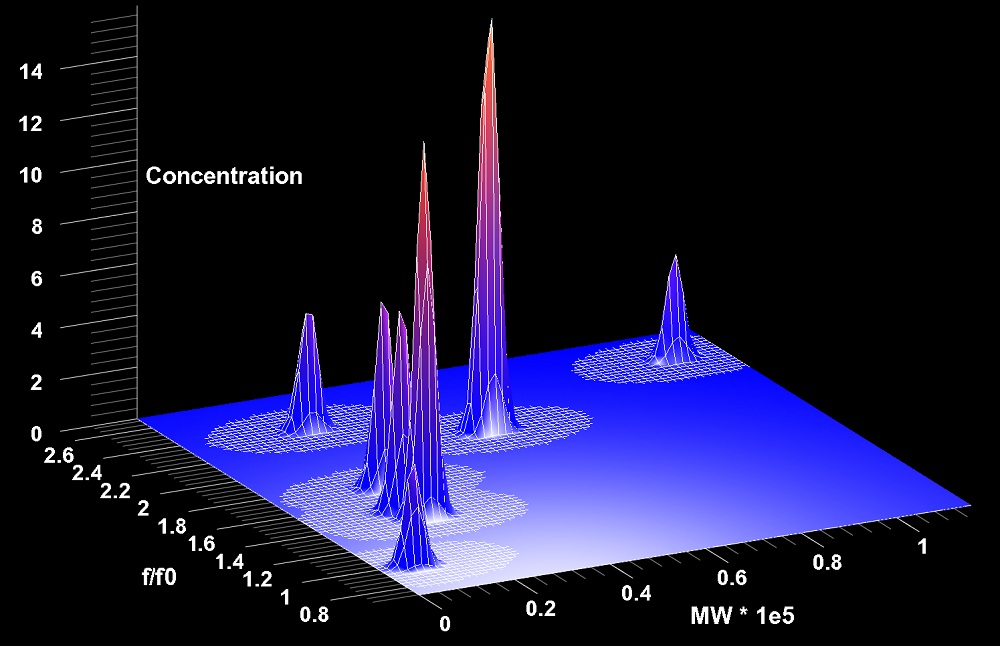
Analytical ultracentrifugation experiments spin samples at very high speeds to study differences in how large molecules such as proteins, DNA and RNA behave in a given solution. In the device different materials diffuse and settle out in different ways, revealing aspects of individual molecules' shapes and sizes. The method has become an essential measurement tool for biochemists.
Analytical ultracentrifugation is also a versatile tool for studying the composition of a mixture of molecules. Even trace amounts of impurities can be resolved using the method, and researchers can analyze mixtures to identify the weights and shapes of each type of molecule in a particular sample.
Computerizing spins
Initially, scientists had to analyze the results of such experiments manually, but with the emergence of computers and sophisticated sensors in the 1960s, researchers developed more precise ways of assessing experimental results.
Today's systems can follow settling and diffusing molecules, detecting how the molecules absorb ultraviolet and visible light, the refractive index of molecules, and how they fluoresce. The systems capture the resulting information digitally, allowing computers to conduct complex analyses.
Get the world’s most fascinating discoveries delivered straight to your inbox.
Borries Demeler, associate professor of biochemistry at the University of Texas Health Sciences Center, serves as the director of the Center for Analytical Ultracentrifugation of Macromolecular Assemblies. For more than two decades, Demeler has worked at the intersection of the physical (spinning samples) and the virtual (supercomputer simulations), investigating new methods and developing software to help researchers make the most of their Analytical ultracentrifugation experiments.
Multitude of experiments
Demeler works with hundreds of investigators around the world, including biophysicists studying the structure and function of biological molecules; materials scientists trying to make more efficient solar cells; and the pharmaceutical industry evaluating the stability of their formulations.
His largest impact, however, is felt through the creation of the UltraScan software package and the development of the UltraScan LIMS portal — a site where researchers can analyze their experimental data over the web using advanced computing methods and systems.
In 2004, Demeler and his colleague, Emre Brookes, began modifying program code so it could run on large-scale computer clusters. That dramatically sped-up the analysis rate for samples. It also enabled the researchers to develop high-resolution analysis methods that address an entirely new class of research questions and widen the applications for the Analytical ultracentrifugation method.
"We often don't know what really is in a [liquid] solution provided by a collaborator, and we need to get the most out of our analysis," Demeler explained. "To fit the data, we simulate many different components that may be in the solution, and ask, 'How much of each component is present in the actual experiment?'"
Computer science
Demeler and his colleagues perform some analyses on a small development cluster in his lab, but for large simulations, the researchers rely on the computing systems of the National Science Foundation-funded Extreme Science and Engineering Discovery Environment, the most powerful and robust collection of integrated, advanced digital resources and services in the world.
Demeler's simulations use up to 14,000 processors simultaneously, speeding up analytic processing by as much as 10,000 times. During a single research phase ending in 2011, Demeler used 3.5 million computing hours on the National Science Foundation-supported Ranger and Lonestar supercomputers at the Texas Advanced Computing Center to perform simulations for the open science community.
"It's not just reserved for biochemists and biophysicists," Demeler said. "We might work with a clinician, perform measurements for materials science or measure the binding strength of a new drug to its target."
Solar particles
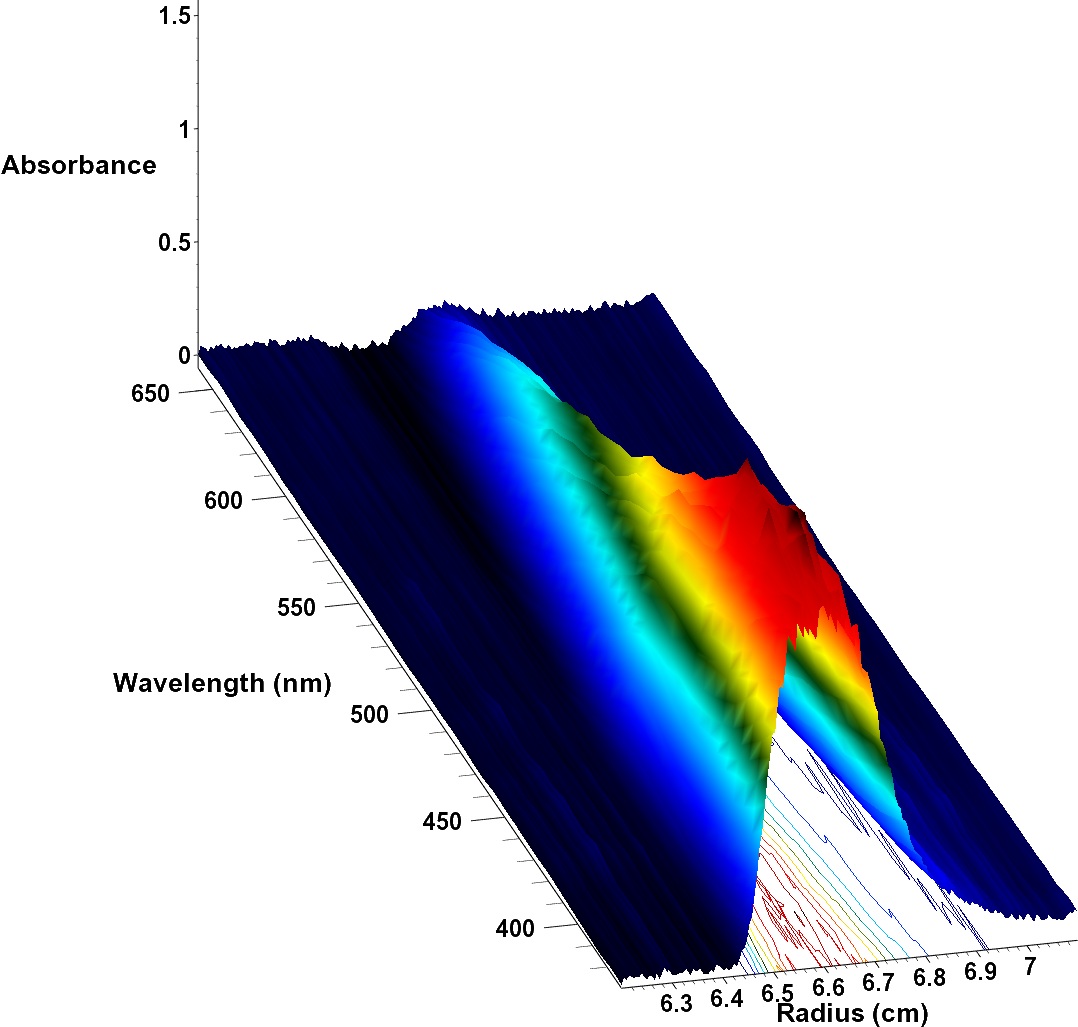
Demeler is collaborating with researchers in Germany to characterize fluorescent nanoparticles made out of cadmium telluride crystals for use in solar panels. Using a new detector developed by collaborators at the Max Planck Institute, he was able to measure hydrodynamic properties of particles, observe their individual absorbance spectra and correlate absorbance properties with particle size.
Whether the application is nanoparticles for industry or biomarkers in blood, analytical ultracentrifugation together with UltraScan is an incredibly powerful methodology. Demeler has made the technology accessible to a broad cohort of lab scientists by developing an easy-to-use web-based gateway.
"The user only has to be familiar with the basic analysis procedure and a web browser, but familiarity with Unix supercomputing is not required," Demeler said. "Our users really like this approach."
Eighty-five-years after its inception, the evolution of the analytic ultracentrifuge continues. Demeler and Brookes' long-term dream is to create a way to integrate all known observational methods, including X-ray crystallography, nuclear magnetic resonance imaging and calorimetry, to see more deeply than current technology allows, without losing sight of the natural conditions under which molecules exist.
"It's like taking a picture of an object from many different angles, and every time you take a picture you see something else that adds to the whole," Demeler said. "By combining them all, the new picture will tell you something you didn't know before."
Editor's Note: The researchers depicted in Behind the Scenes articles have been supported by the National Science Foundation, the federal agency charged with funding basic research and education across all fields of science and engineering. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author and do not necessarily reflect the views of the National Science Foundation. See the Behind the Scenes Archive.
 Live Science Plus
Live Science Plus