1st draft of a human 'pangenome' published, adding millions of 'building blocks' to the human reference genome
A new version of the human reference genome incorporates genetic data from 47 individuals from around the globe, deepening scientists' view into how genes work.

Get the world’s most fascinating discoveries delivered straight to your inbox.
You are now subscribed
Your newsletter sign-up was successful
Want to add more newsletters?

Delivered Daily
Daily Newsletter
Sign up for the latest discoveries, groundbreaking research and fascinating breakthroughs that impact you and the wider world direct to your inbox.

Once a week
Life's Little Mysteries
Feed your curiosity with an exclusive mystery every week, solved with science and delivered direct to your inbox before it's seen anywhere else.

Once a week
How It Works
Sign up to our free science & technology newsletter for your weekly fix of fascinating articles, quick quizzes, amazing images, and more

Delivered daily
Space.com Newsletter
Breaking space news, the latest updates on rocket launches, skywatching events and more!

Once a month
Watch This Space
Sign up to our monthly entertainment newsletter to keep up with all our coverage of the latest sci-fi and space movies, tv shows, games and books.

Once a week
Night Sky This Week
Discover this week's must-see night sky events, moon phases, and stunning astrophotos. Sign up for our skywatching newsletter and explore the universe with us!
Join the club
Get full access to premium articles, exclusive features and a growing list of member rewards.
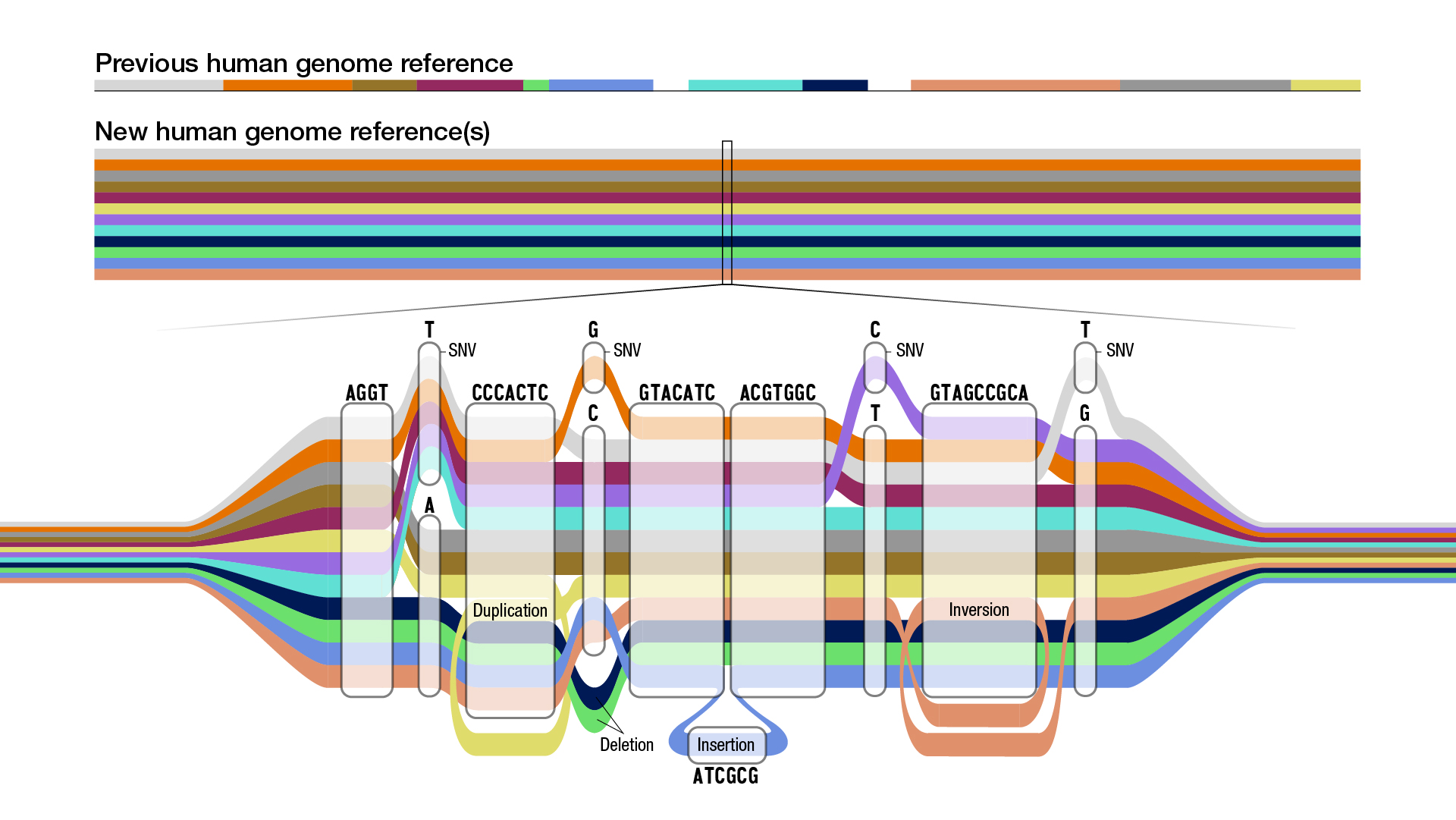
Scientists have published the first human "pangenome" — a full genetic sequence that incorporates genomes from not just one individual, but 47.
These 47 individuals hail from around the globe and thus vastly increase the diversity of the genomes represented in the sequence, compared to the previous full human genome sequence that scientists use as their reference for study. The first human genome sequence was released with some gaps in 2003 and only made "gapless" in 2022. If that first human genome is a simple linear string of genetic code, the new pangenome is a series of branching paths.
The ultimate goal of the Human Pangenome Reference Consortium, which published the first draft of the pangenome on Wednesday (May 10) in the journal Nature, is to sequence at least 350 individuals from different populations around the world. Although 99.9% of the genome is the same from person to person, there is a lot of diversity found in that final 0.1%.
"Rather than using a single genome sequence as our coordinate system, we should instead have a representation that is based on the genomes of many different people so we can better capture genetic diversity in humans," Melissa Gymrek, a genetics researcher at the University of California, San Diego, who was not involved in the project, told Live Science.
Related: More than 150 'made-from-scratch' genes are in the human genome. 2 are totally unique to us.
A reference for health
The first full human genome sequence was completed in 2003 by the Human Genome Project and was based on one person's DNA. Later, bits and pieces from about 20 other individuals were added, but 70% of the sequence scientists use to benchmark genetic variation still comes from a single person.
Geneticists use the reference genome as a guide when sequencing pieces of people's genetic codes, Arya Massarat, a doctoral student in Gymrek's lab who co-authored an editorial about the new research with her in the journal Nature, told Live Science. They match the newly decoded DNA snippets to the reference to figure out how they fit within the genome as a whole. They also use the reference genome as a standard to pinpoint genetic variations — different versions of genes that diverge from the reference — that might be linked with health conditions.
Get the world’s most fascinating discoveries delivered straight to your inbox.
But with a single reference mostly from one person, scientists have only a limited window of genetic diversity to study.
The first pangenome draft now doubles the number of large genome variants, known as structural variants, that scientists can detect, bringing them up to 18,000. These are places in the genome where large chunks have been deleted, inserted or rearranged. The new draft also adds 119 million new base pairs, meaning the paired "letters" that make up the DNA sequence, and 1,115 new gene duplication mutations to the previous version of the human genome.
"It really is understanding and cataloging these differences between genomes that allow us to understand how cells operate and their biology and how they function, as well as understanding genetic differences and how they contribute to understanding human disease," study co-author Karen Miga, a geneticist at the University of California, Santa Cruz, said at a press conference held May 9.
The pangenome could help scientists get a better grasp of complex conditions in which genes play an influential role, such as autism, schizophrenia, immune disorders and coronary heart disease, researchers involved with the study said at the press conference.
For example, the Lipoprotein A gene is known to be one of the biggest risk factors for coronary heart disease in African Americans, but the specific genetic changes involved are complex and poorly understood, study co-author Evan Eichler, a genomics researcher at the University of Washington in Seattle, told reporters. With the pangenome, researchers can now more thoroughly compare the variation in people with heart disease and without, and this could help clarify individuals' risk of heart disease based on what variants of the gene they carry.
Related: As little as 1.5% of our genome is 'uniquely human'
A diverse understanding
The current pangenome draft used data from participants in the 1000 Genomes Project, which was the first attempt to sequence genomes from a large number of people from around the world. The included participants had agreed for their genetic sequences to be anonymized and included in publicly available databases.
The new study also used advanced sequencing technology called "long-read sequencing," as opposed to the short-read sequencing that came before. Short-read sequencing is what happens when you send your DNA to a company like 23andMe, Eichler said. Researchers read out small segments of DNA and then stitch them together into a whole. This kind of sequencing can capture a decent amount of genetic variation, but there can be poor overlap between each DNA fragment. Long-read sequencing, on the other hand, captures big segments of DNA all at once.
While it's possible to sequence a genome with short-read sequencing for about $500, long-read sequencing is still expensive, costing about $10,000 a genome, Eichler said. The price is coming down, however, and the pangenome team hopes to sequence their next batches of genomes at half that cost or less.
The researchers are working to recruit new participants to continue to fill in diversity gaps in the pangenome, study co-author Eimear Kenny, a professor of medicine and genetics at the Institute for Genomic Health at Icahn School of Medicine at Mount Sinai in New York City, told reporters. Because genetic information is sensitive and because different rules govern data-sharing and privacy in different countries, this is delicate work. Issues include privacy, informed consent, and the possibility of discrimination based on genetic information, Kenny said.
Already, researchers are uncovering new genetic processes with the draft pangenome. In two papers published in Nature alongside the work, researchers looked at highly repetitive segments of the genome. These segments have traditionally been difficult to study, biochemist Brian McStay of the National University of Ireland Galway, told Live Science, because sequencing them via short-read technology makes it hard to understand how they fit together. The long read technology allows for long chunks of these repetitive sequences to be read at once.
The studies found that in one type of repetitive sequence, known as segmental duplications, there is a larger than expected amount of variation, potentially a mechanism for the long-term evolution of new functions for genes. In another type of repetitive sequence that is responsible for building the cellular machines that create new proteins, though, the genome stays remarkably stable. The pangenome allowed researchers to discover a potential mechanism for how these key segments of DNA stay consistent over time.
"This is just the start," McStay said. "There will be a whole lot of new biology that will come out of this."

Stephanie Pappas is a contributing writer for Live Science, covering topics ranging from geoscience to archaeology to the human brain and behavior. She was previously a senior writer for Live Science but is now a freelancer based in Denver, Colorado, and regularly contributes to Scientific American and The Monitor, the monthly magazine of the American Psychological Association. Stephanie received a bachelor's degree in psychology from the University of South Carolina and a graduate certificate in science communication from the University of California, Santa Cruz.
 Live Science Plus
Live Science Plus











