Secret inner workings of cells revealed through self-assembling 'memory' chains
Researchers genetically altered mouse brain cells to produce physical timelines of their key events in the form of a self-assembling chain of fluorescent proteins.
Researchers have coaxed mouse brain cells into producing self-assembling protein chains that can record information, or "memories," about the hidden processes that take place within the cells. Once fully formed, these biological black boxes can be easily read using a light microscope, which could potentially revolutionize how scientists study cellular processes and the diseases that affect them.
Cells are hubs of constant activity, carrying out the crucial everyday tasks that keep organisms alive. This activity is coordinated by specific "cellular events," such as the expression of certain genes or the triggering of cellular pathways, a series of interactions between molecules in a cell that leads to a certain product or a change in a cell. But understanding exactly how these cellular events play out can be challenging.
By imaging the proteins, RNA or other molecules created during these events inside the cells, scientists have learned how most cellular events work. However, this method provides only a brief snapshot of the event. And, although these snapshots can be stitched together to form a loose picture, scientists likely miss a lot of what is really going on.
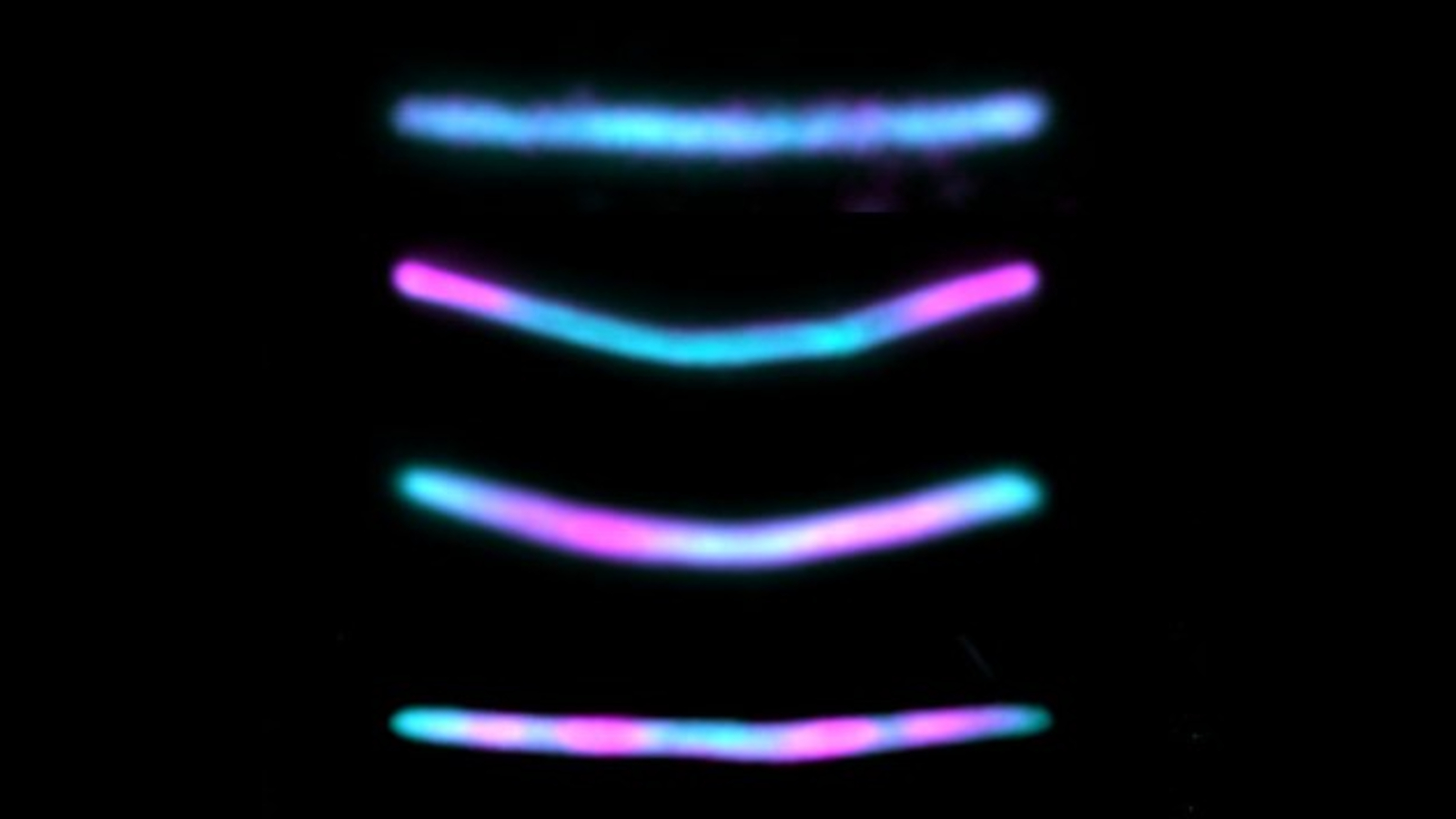
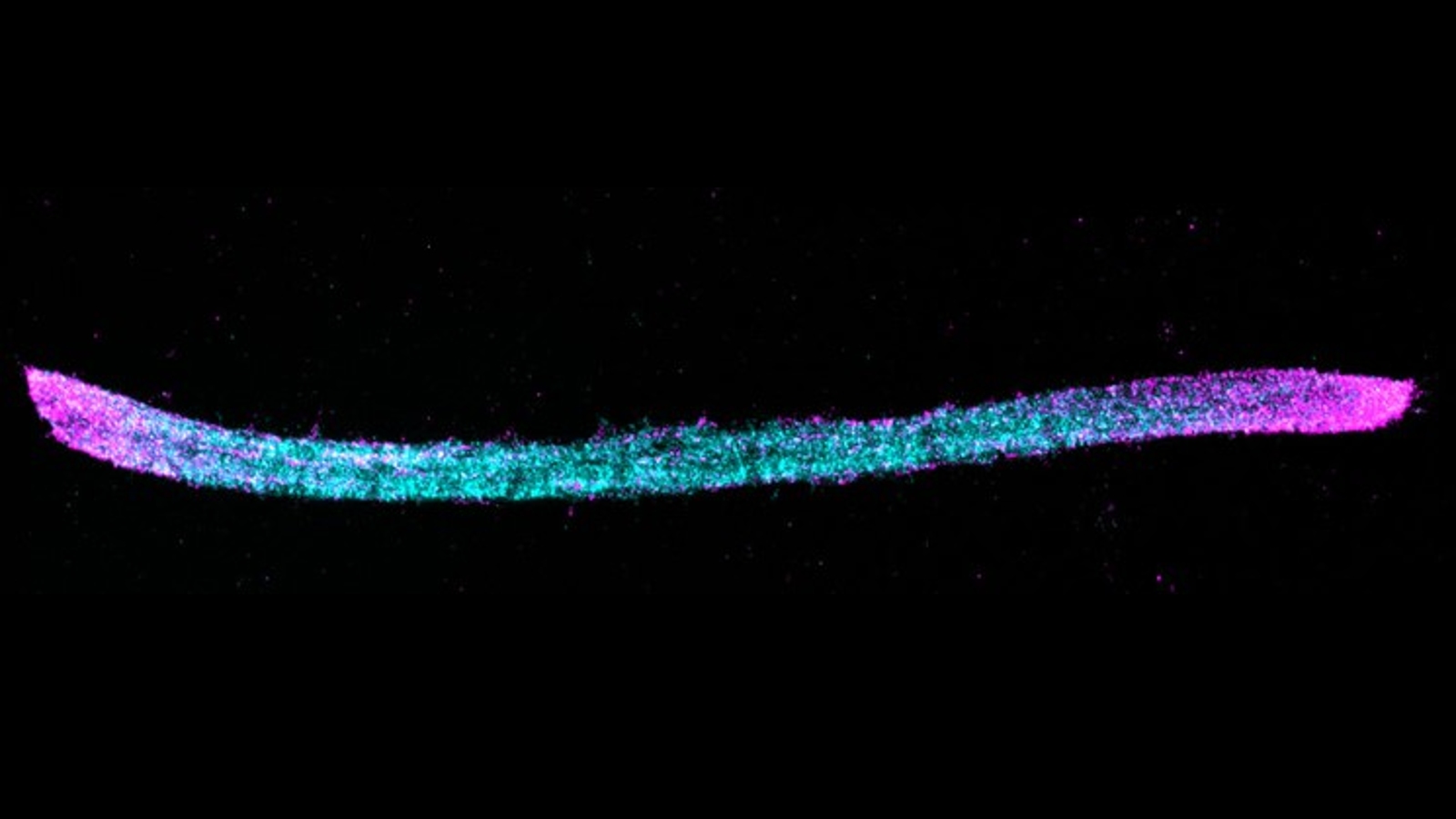
In a new study, published Jan. 2 in the journal Nature Biotechnology, researchers genetically altered mouse neurons to create physical timelines of these events. The hacked brain cells continually produced identical fluorescent protein subunits, which naturally self-assembled into a long chain. When important cellular events — such as a specific gene being turned on — occurred, an alternate subunit was produced by the cells instead and was added into the chain in place of the normal recurring subunit. This allowed the researchers to go back and look at the chains to see exactly when these cellular events happened.
"It's not only a snapshot in time, but also records past history," study lead author Changyang Linghu, a cell biologist at University of Michigan, said in a statement. "Just like how tree rings can permanently store information over time as the wood grows."
Related: Synthetic brain cells that store 'memories' are possible, new model reveals
During the new experiments, researchers grew cultures of the genetically altered mouse neurons in petri dishes. The hacked brain cells were capable of producing two protein subunits: HA, which was continually produced by the cell, and V5, which was produced instead of HA every time a gene called c-Fos — which is activated in neurons as memories are formed in mice and humans — was switched on.
Get the world’s most fascinating discoveries delivered straight to your inbox.
Each of the two subunits, which are not produced by normal mouse neurons, had a uniquely colored fluorescent antibody attached via a short peptide known as an epitope tag, which makes it easy to differentiate under a microscope. The HA subunit had a blue tag, and the V5 antibody had a pink tag. The resulting chains therefore looked like long, blue lines, with the occasional pink section sprinkled in every time the c-Fos gene was activated. This allowed the researchers to count how often the c-Fos gene was activated and how much time passed between each activation.
In principle, if the same method were applied to neurons from humans, it could allow researchers to see how and when people form new memories, which could be used to help study neurological conditions such as dementia. However, this study is only a proof-of-concept and it will be years, if not decades, before the protein chains can be used in a clinical setting.
In addition, the team believes that this method could eventually be used in any type of cell to create timelines of when multiple different genes are activated. Additional subunits also could be produced for other cellular events, potentially revealing the hidden inner workings of almost any cell type and how they interact with one another, which could be a game-changer in medicine, the researchers said.
However, there is one major limitation to the memory chains: They can grow only as long as the cell is wide. Once the chain hits the inside of the cell wall, there is nowhere left for it to go and it will start to become tangled and unreadable.
During the experiments, the researchers created memory chains over the space of around two days before they hit the cell wall. Microscope images were taken just before this happened to preserve the data.
In theory, the rate at which the subunits are added to the chains can be reduced so that the end chain is still the same length but takes longer to form, which, in turn, could allow the researchers to record more specific events, Linghu said. But doing this would reduce the accuracy of the timeline because there would be more uncertainty over exactly when the event occurred, he added.

Harry is a U.K.-based senior staff writer at Live Science. He studied marine biology at the University of Exeter before training to become a journalist. He covers a wide range of topics including space exploration, planetary science, space weather, climate change, animal behavior and paleontology. His recent work on the solar maximum won "best space submission" at the 2024 Aerospace Media Awards and was shortlisted in the "top scoop" category at the NCTJ Awards for Excellence in 2023. He also writes Live Science's weekly Earth from space series.
 Live Science Plus
Live Science Plus