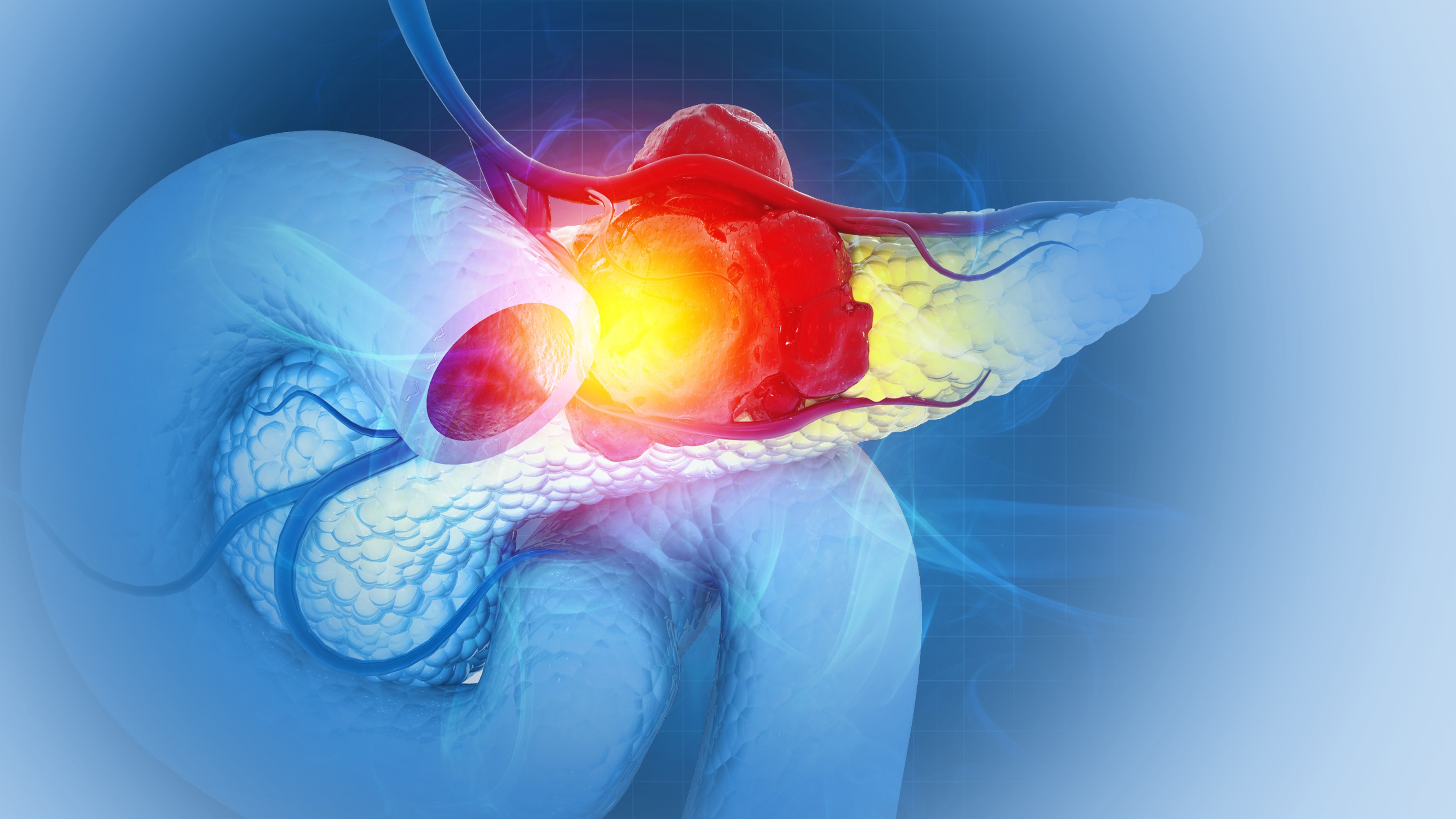
DeepMind's AI program AlphaFold3 can predict the structure of every protein in the universe — and show how they function
AlphaFold3 uses AI to helps scientists more accurately predict how proteins interact with other biological molecules.
DeepMind has unveiled the third version of its artificial intelligence (AI)-powered structural biology software, AlphaFold, which models how proteins fold.
Structural biology is the molecular basis study of biological materials — including proteins and nucleic acids —and aims to reveal how they are structured, work, and interact.
AlphaFold3 helps scientists more accurately predict how proteins — large molecules that play a critical role in all life forms, from plants and animals to human cells — interact with other biological molecules, including DNA and RNA. Doing so will enable scientists to “truly understand life’s processes,” DeepMind representatives wrote in a blog post.
By comparison, its predecessors, AlphaFold and AlphaFold2, could only predict the shapes that proteins fold into. That was still a major scientific breakthrough at the time.
AlphaFold3's predictions could help scientists develop bio-renewable materials, crops with greater resistance, new drugs and more, the research team wrote in a study published May 8 in the journal Nature.
Given a list of molecules, the AI program can show how they fit together. It does this not only for large molecules like proteins, DNA, and RNA but also for small molecules known as ligands, which bind to receptors on large proteins like key fitting into a lock.
Get the world’s most fascinating discoveries delivered straight to your inbox.
AlphaFold3 also models how some of these biomolecules (organic molecules produced by living things) are chemically modified. Disruptions in these chemical modifications can play a role in diseases, according to the blog post.
AlphaFold3 can perform these calculations because its underlying machine-learning architecture and training data encompasses every type of biomolecule.
The researchers claim that AlphaFold3 is 50% more accurate than current software-based methods of predicting protein structures and their interactions with other molecules.
For example, in drug discovery, Nature reported that AlphaFold3 outperformed two docking programs — which researchers use to model the affinity of small molecules and proteins when they bind together — and RoseTTAFold All-Atom, a neural network for predicting biomolecular structures.
Frank Uhlmann, a biochemist at the Francis Crick Institute in London, told Nature that he's been using the tool for predicting the structure of proteins that interact with DNA when copying genomes and experiments show the predictions are mostly accurate.
However, unlike its predecessors, AlphaFold 3 is no longer open source. This means scientists cannot use custom versions of the AI model, or access its code or training data publicly, for their research work.
Scientists looking to use AlphaFold3 for non-commercial research can access it for free via the recently launched AlphaFold Server. They can input their desired molecular sequences and gain predictions within minutes. But they can only perform 20 jobs per day.
Nicholas Fearn is a freelance technology and business journalist from the Welsh Valleys. With a career spanning nearly a decade, he has written for major outlets such as Forbes, Financial Times, The Guardian, The Independent, The Daily Telegraph, Business Insider, and HuffPost, in addition to tech publications like Gizmodo, TechRadar, Computer Weekly, Computing and ITPro.
 Live Science Plus
Live Science Plus