Person who had measles 100 years ago helps scientists trace origins of virus
A preserved lung helped scientists rewrite the virus's history.
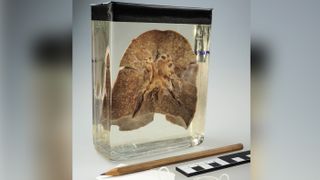
A diseased human lung, fixed in the preservative formalin for more than 100 years, helped scientists trace the history of the measles virus and place its origin as far back as the sixth century B.C.
For years, the lung sat in the basement of the Berlin Museum of Medical History along with hundreds of other lung specimens, all collected and preserved between the 1870s and 1930s. On a hunt for well-preserved respiratory pathogens, virologist Sébastien Calvignac-Spencer of the Robert Koch Institute and his research team descended into the basement and peered into each and every jar. "It's a matter of serendipity" that the team found a lung belonging to a 2-year-old measles patient who died from the disease in 1912, Calvignac-Spencer said.
The team managed to extract samples of the virus from the 108-year-old lung tissue and used the genetic material — the oldest measles genome ever sequenced — to learn more about the origins of the pathogen. In a new study, published today (June 18) in the journal Science, they estimate that measles could have diverged from its closest known relative, a now eradicated cattle virus, as early as 528 B.C.
The new estimate suggests that the virus may be "more than a 1,000 years older than any previous estimate," Calvignac-Spencer told Live Science.
Related: 11 (sometimes) deadly diseases that hopped across species
A rare find
Previous studies predicted that the measles and the extinct cattle virus, called rinderpest, split from their most recent common ancestor between the 11th and 12th centuries, according to a 2011 report in the journal Molecular Biology and Evolution (MBE). However, the Persian physician Muhammad ibn Zakariya al-Razi wrote a clinical description of measles in the 10th century, so something didn't add up.
"The split between measles and rinderpest is clearly underestimated," said Joel Wertheim, author of the MBE report and assistant professor of medicine at the University of California, San Diego, who was not involved in the new Science study. These underestimates arise from two critical issues: a lack of old measles samples and faulty assumptions about how the virus mutates through time, which skew evolutionary models toward a "ridiculously recent date," Wertheim told Live Science.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
Wertheim and his coauthors built a new model to account for these factors and pushed back the origin date to the end of the ninth century, but "we didn't think we were right," he said. Now, Calvignac-Spencer and his team have reached a more realistic estimate, in part, by including the newly-discovered 1912 specimen in their analysis, Wertheim said.
Related: Going viral: 6 new findings about viruses

Before the team found the 1912 sample, the oldest measles genome ever sequenced dated to 1954, the authors noted. Scientists estimate the rate of evolutionary change, or how much and how quickly a virus mutates, by comparing samples gathered at different times and tracking differences in their genetic code. The more and older samples we examine, the clearer that rate of change becomes, Calvignac-Spencer said.
But the backbone of the measles virus is RNA, a type of genetic material that degrades quickly compared with its sturdier cousin DNA. The 1912 sample escaped this fate because the lung had been fixed in formalin, a preservative that halts chemical reactions that would otherwise degrade RNA. Formalin also "glues" the preserved RNA to nearby molecules, making it difficult to extract, Calvignac-Spencer said.
To unstick the RNA, the team sliced 0.007 ounces (200 milligrams) of tissue from the lung and boiled the tiny sample, causing the sticky molecules within to come apart without destroying the RNA. The team then constructed "an almost complete" genome from the rescued RNA, they wrote. To further enrich their evolutionary model, the team scoured the collection of genetic samples in the German National Reference Laboratory and found two measles samples collected in 1960 to add to their analysis.
Building better models
The team built their evolutionary model from the 1912 sample, 1960 samples and 127 additional samples, most collected in or after the 1990s. A second model compared about 50 measles sequences to the rinderpest virus, which was declared eradicated in 2011, and its closest relative peste des petits ruminants (PPRV), which infects goats and sheep, to pin down when these pathogens split from their shared ancestor.
In both of these models, the team took a phenomenon called "purifying selection" into account, which many previous studies overlooked, Calvignac-Spencer said. While some evolutionary pressures add helpful mutations to the genome, and keep it stable over time, so-called purifying selection purges harmful mutations from the genome before they can accumulate. These complementary forces help set the pace of evolutionary change, so to estimate when measles first emerged, you must factor in purifying selection, Wertheim said.
"You can change [your estimates] by an order of magnitude by taking purifying selection into account," he said. Purifying selection, in part, causes certain segments of the genome to mutate readily and often, while others hardly change at all, he added. "You will have several mutations hitting the same position over and over," but since you only have a limited number of samples, you may miss some of those mutations, Calvignac-Spencer said. The team designed their model to capture these mutations that may otherwise be missed.
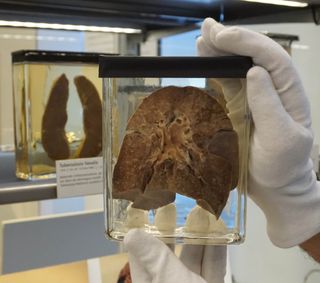
Based on when rinderpest and measles diverged, the "earliest possible date for the establishment of measles in human populations" occurred around the sixth century B.C., although the exact date that the virus first infected people remains unknown.
The authors noted that, around 2,000 to 2,500 years ago, humans began building settlements large enough to sustain a measles outbreak, offering the virus an opportunity to establish itself. Measles tends to peter out in communities of fewer than 250,000 individuals, as residents quickly become immune to, or die from, the disease, so "small human populations could only serve as dead-end hosts," they wrote.
Calvignac-Spencer said that he's interested in uncovering ancient measles samples, if they exist, in order to further refine our understanding of the pathogen's history. Wertheim said that he predicts more virologists will join the hunt for ancient samples lurking in museum basements and hospital archives.
"I was astonished when I saw they were able to pull out an over 100-year-old virus from lung tissue," Wertheim said. I think that more virologists will begin using "older and older viruses as people become more ambitious and encouraged by these results," he added.
- 20 of the worst epidemics and pandemics in history
- The 12 deadliest viruses on Earth
- Top 10 mysterious diseases
Originally published on Live Science.
<a href="https://www.livescience.com/download-your-favorite-magazines.html" data-link-merchant="livescience.com"" target="_blank">OFFER: Save 45% on 'How It Works' 'All About Space' and 'All About History'!
For a limited time, you can take out a digital subscription to any of <a href="https://www.livescience.com/download-your-favorite-magazines.html" data-link-merchant="livescience.com"" data-link-merchant="livescience.com"" target="_blank">our best-selling science magazines for just $2.38 per month, or 45% off the standard price for the first three months.

Nicoletta Lanese is the health channel editor at Live Science and was previously a news editor and staff writer at the site. She holds a graduate certificate in science communication from UC Santa Cruz and degrees in neuroscience and dance from the University of Florida. Her work has appeared in The Scientist, Science News, the Mercury News, Mongabay and Stanford Medicine Magazine, among other outlets. Based in NYC, she also remains heavily involved in dance and performs in local choreographers' work.
Most Popular



