How Bacteria Get Past Our Defenses
This Behind the Scenes article was provided to LiveScience in partnership with the National Science Foundation.
Mucus is more than gross — it's a critical barrier against disease, trapping many of the germs that want to invade your body. A wet mesh of proteins, antiseptic enzymes and salts, mucus is what keeps all but a few microbes from wreaking havoc on many of our most exposed tissues.
Helicobacter pylori is one of the few. The tiny, corkscrew-shaped microbe bores through the mucus that lines the acidic cauldron of the human stomach, establishing colonies on the cells below.
After invading the stomach lining, H. pylori causes persistent, low-grade irritation that over time can lead to ulcers, and if untreated, to cancer.
Boston University (BU) physicist Rama Bansil — along with students and colleagues from BU, Harvard Medical School and MIT — recently helped discover how H. pylori gets through our defenses. The findings could help us protect against this germ, as well as many others.
For decades, Bansil has been studying the physics of gels, and since 1990, a gel composed primarily of mucin, the glyco-protein (protein and sugar complex) found in mucus.
"The mucins of different organs are similar overall, but they have slightly different structures and properties depending on where they are found in the body," Bansil said. "Some become gel, others don’t. They’re tuned to their function. It’s in fish, it’s in slugs — slugs use it to move.” In fact, all vertebrates produce mucin, and many human diseases involve the material.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
Bansil’s studies have become so associated with mucin — particularly of the stomach —some of her colleagues refer to her laboratory as the Snot Research Lab.
"In some ways, I think my getting into stomach research was serendipity," Bansil said. Nearly twenty years ago, colleagues approached her looking for a gel expert, an addition to an interdisciplinary team for studying the mucus in our digestive system.
As the researchers started delving deeper into the research problems, they realized that they needed more collaborators and techniques to help find answers.
"You can’t just work with crude mucus," Bansil said. "For stomach mucus, purifying it to obtain the active ingredient, the mucin, is a laborious task. That may be why there are very few groups studying the biophysics of mucin. Protein chemistry is a huge field, but the study of mucin itself is not as advanced — it’s a very complicated protein." In fact, many of the leading studies on mucin were conducted abroad in Europe.
"Originally, our team was just a few collaborators at the BU school of medicine," said Bansil. The medical part of the group later moved to Harvard Medical School, and now the team also includes researchers at MIT. (The full team is listed in a recent press release).
"I would tell colleagues that we were looking at this interesting problem and I was giving a lot of talks about why the stomach doesn’t digest itself, and this helped recruit colleagues. The first person I cornered was the person in the lab next to me; we collaborated on atomic force microscopy."
The microscopy allowed the research team to see mucus up close, and revealed the structure of single mucin molecules.
After several years of working out the basic physical properties of mucin and how those proteins protect against acid in the stomach, the research team wanted to pursue mucin’s relationships to disease.
It was in 1993 — when Bansil came across an article in the New Yorker on the link between H. pylori and ulcers — that she decided to tackle the mystery of how H. pylori travels through stomach mucus. However, it took more than ten years before the researchers actually started working with bacteria.
H. pylori has been a popular subject for study in recent years, particularly following the research of pathologist Robin Warren and clinical researcher Barry Marshall, both of Perth, Western Australia, in the early 1980s. Warren and Marshall definitively linked the bacteria to the stomach, and to ulcers, overturning the persistent belief that bacteria could not thrive in such an acidic environment. Ultimately, the two researchers won the 2005 Nobel Prize in Medicine for their efforts.
Many researchers have further studied H. pylori, learning more about its structure, how it thrives, and even how it fends off stomach acid. Yet, until now no one had explored how it traveled through the sticky gels of stomach mucus.
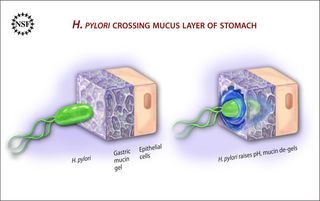
Conventional wisdom held that corkscrew-shaped H. pylori relies on its shape to twist and bore its way through mucus.
Instead, as part of the thesis of BU doctoral student Jonathan Celli, the researchers found that the bacteria swim in a manner more like other bacteria with whip-like tails, H. pylori just changes its environment to make movement possible.
"We figured out that that it doesn’t move like a corkscrew — everyone thought it did…and the same biochemistry that it uses for survival makes it possible for it to move," explained Bansil. "These two functions are inextricably coupled. It’s chemically affecting its environment, and then it basically acts like a snowplow, moving by altering its surroundings."
H. pylori secretes the enzyme urease, which interacts with urea in the stomach to produce ammonia — the ammonia is what neutralizes the acids in the immediate environment. The less-acidic environment de-gels the mucin, allowing the microbe to travel through it using standard, flagella-based locomotion, much like other swimming bacteria.
To confirm their findings, the researchers placed H. pylori into an acidic mucin gel in a laboratory setting. While its flagella moved, the organism could not. After the microbes secreted urease and acidity diminished, the microbes were able to forge through the gel.
Bansil and her colleagues next want to understand the progress of H. pylori-related diseases, particularly in the context of living hosts. The team is planning to work on new imaging techniques that may reveal even greater detail about the organisms and how they inflict damage on the human body.
Jonathan Celli, supported by an NSF GK-12 fellowship, was lead author on the H. pylori findings published in the Proceedings of the National Academy of Sciences on Aug. 11, 2009.
- Video - Flapping Bacterial Flagella
- Fake Snot Makes Electronic Nose Smell Better
- Bacteria Thrive in Hostile Human Bellies
Editor's Note: This research was supported by the National Science Foundation (NSF), the federal agency charged with funding basic research and education across all fields of science and engineering. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author and do not necessarily reflect the views of the National Science Foundation. See the Behind the Scenes Archive.
Most Popular



